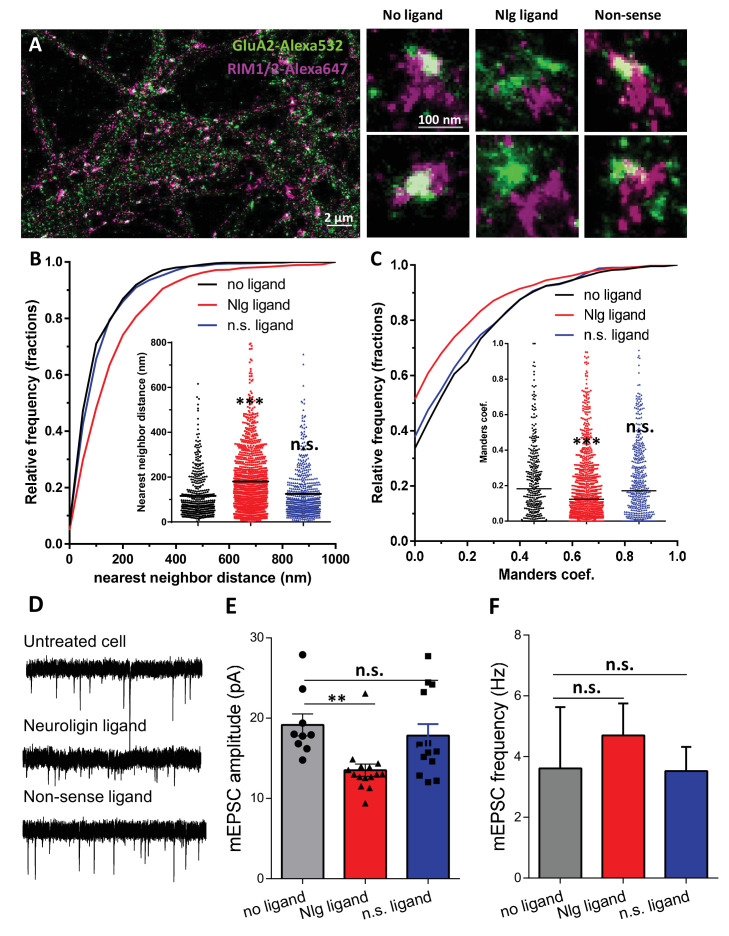
Figure 5. Acute disruption of PSD95-NLG interaction impaired both pre-post alignment and synaptic transmission.
(A) Example of dual-color d-STORM image of GluA2 containing AMPAR labelled with Alexa 532 nm and RIM labelled with Alexa 647 nm. Right panels, examples of GluA2 and RIM co-labeling without ligand or after 1-day treatment with Nlg biomimetic ligand or non-sense ligand (from the left to the right). (B and C) presents the quantification of this co-localization. (B) Cumulative distribution of the bivariate nearest neighbor distance between GluA2 and RIM clusters without ligand (black), with NLG ligand NLG1 (green) or non-sense ligand (blue). These data demonstrate a loss of the pre-post synaptic alignment in the presence of NLG ligand. (C) Manders’ coefficients calculated between GluA2 nanodomains and RIM clusters in function of ligand treatment (n = 4; 5 and 4 Control, NLG ligand and non-sense ligand respectively, corresponding to 451; 1311 and 640 independent pairs of co-localization). (D) Example of mEPSC traces recorded in cultured neurons without ligand, with NLG ligand or with non-sense ligand. (E and F) average of the mEPSC amplitude and amplitude recorded when neurons in culture are incubated without ligand (dark) or with either NLG ligand (red) or with non-sense ligand (blue) (n = 9; 15 and 13 respectively). mEPSCs amplitude is decreased by 30% in neurons incubated with NLG ligand.