Figure 3. Effect of the c.301+2T>A change on transcript processing and protein stability.
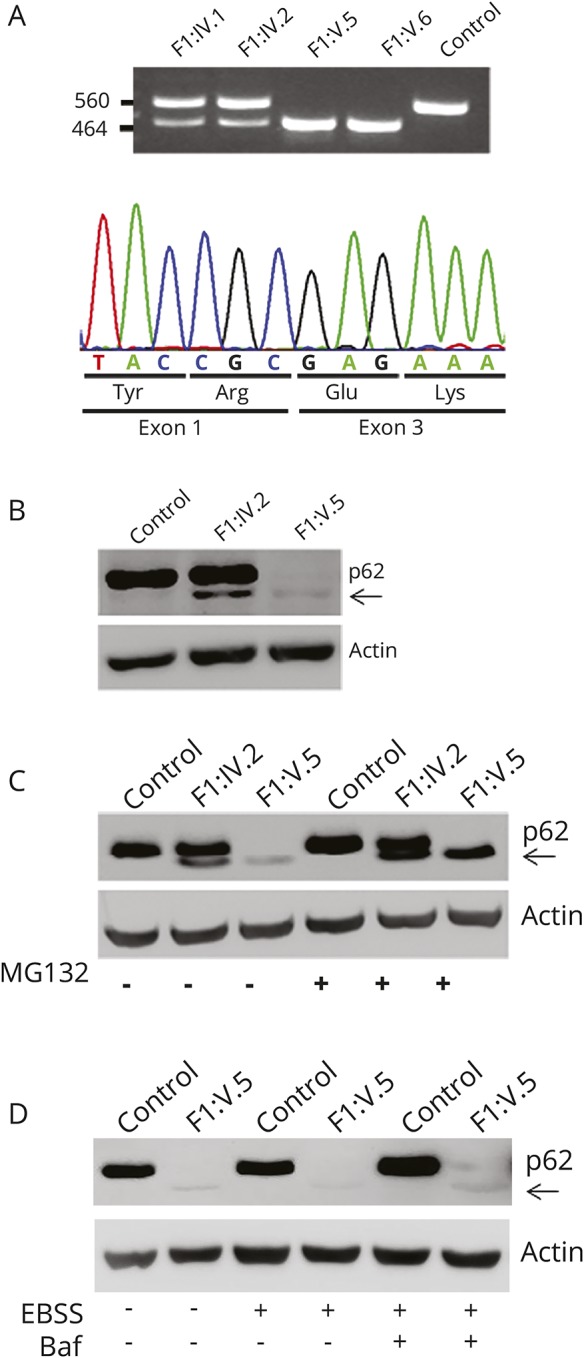
(A) The homozygous c.301+2T>A substitution causes skipping of coding exon 2. Amplification of the cDNA fragment encompassing coding exons 1 to 4 reveals an aberrant PCR product lacking exon 2 in the 2 affected siblings; wild-type and aberrant processed transcripts are observed in both parents (top). Sequencing of the mature SQSTM1 transcript from affected individual F1:V.5 documents skipping of coding exon 2 and the preservation of the phase of the reading frame (bottom). (B) Western blot analysis performed on skin fibroblasts from individual F1:V.5 (right), the unaffected mother (middle), and an unrelated individual (left), indicating that the mutated SQSTM1 protein lacking the amino acid portion encoded by exon 2 (arrow) has a dramatically reduced level compared to the wild-type protein. (C) Treatment of fibroblasts with MG132 (6 hours), an inhibitor of proteasome function, results in a marked increased level of the SQSTM1 mutant, indicating that this protein is rapidly degraded via proteasome. (D) Incubation with bafilomycin A1 (baf) and Earle balanced salt solution (EBSS) (6 hours) does not result in any significant change in the level of the mutated SQSTM1 protein, ruling out a major role of autophagy in the degradation of the SQSTM1 mutant.
