Figure 4.
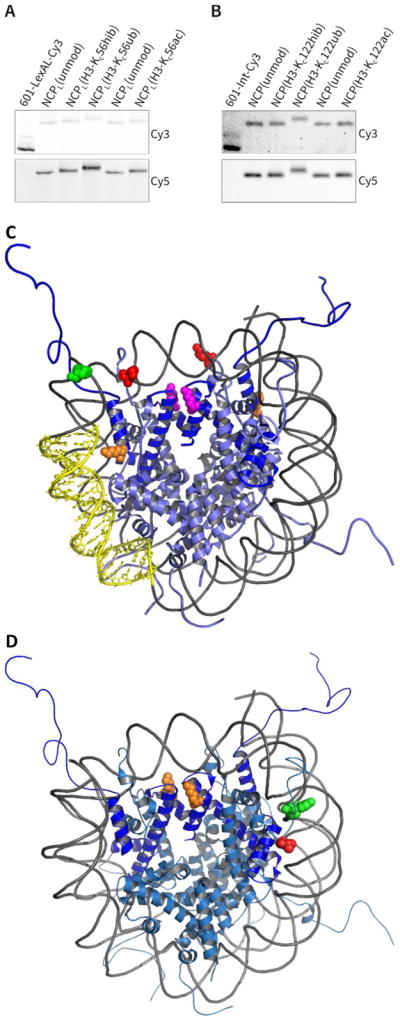
Native PAGE of reconstituted nucleosomes bearing hydrazide mimics (NCP stands for nucleosome core particle). Two types of nucleosomes we prepared to study their dynamics: NCPL with LexA target sequence in the dsDNA and standard NCP which does not include LexA dsDNA. (A) Nucleosomes with H3-Kc56 modifications. (B) Nucleosomes containing H3-Kc122 modifications. (C and D) Structures of NCPs (PDB ID:22 1KX5), depicting the location of fluorophores and PTM mimics. Both copies of H3 are rendered in blue. Locations of Cy3 (green), Cy5 (red), and PTM mimics are in shown as ‘spheres’. (C) Nucleosomes used in the LexA binding studies show LexA target sequence (yellow ‘sticks’), H3-K56 (orange), and H3-K122 (magenta). (D) NCPs used in salt titrations and kinetics show H3-K122 residues in orange. H3-unmod (as described in Figure 3 legend above) was used to assemble both NCPL(unmod) and NCP(unmod) in Figures 4A and 4B, respectively. Also, these NCPs were loaded on every third lane to help reveal the gel shifts due to PTMs. This observation has been noted in previous reports (see Fig. 3c–d in reference 5d or Fig. 2A in reference 21c).
