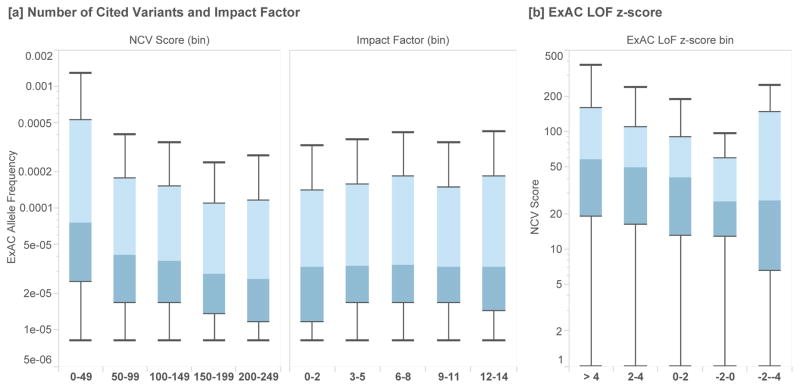
Figure 1.
[a] Box plots of the allele frequency (Exome Aggregation Consortium, N=60,706) of a set of putative disease variants (HGMD 2012.1), grouped by the number of cited variants (NCV) in each gene and the journal impact factor for each citation. We observe an inverse correlation between the NCV score and allele frequency, while the impact factor of the journal in which a variant is cited is uncorrelated with allele frequency. [b] We find that genes under stronger selective constraint (those with positive z-scores reflect genes that have fewer loss of function [LoF] variants than expected based on a mutational model) have higher NCV scores than those genes which are already associated with disease but under lower selective constraint (p-value = 2.36×10−3).