Figure 2.
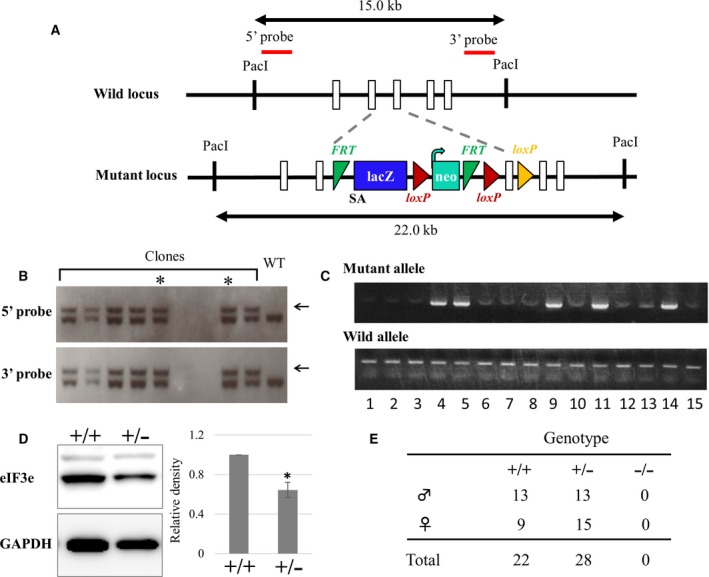
Targeted disruption of mouse eIF3e. (A) Scheme for generating eIF3e‐deficient mice. The genomic structure of eIF3e and the desired homologous recombination allele along with southern blotting probes are shown. Open boxes represent eIF3e exons, and the other functional sequences or cassettes are illustrated with their names. SA refers to the splicing acceptor for expressing lacZ and that modifies the mRNA structure of eIF3e for target disruption. Dotted lines show the insertion region that results in desired recombination. Numerical value (kb) with double arrows indicates genomic fragment size digested with PacI. (B) Representative results of southern blotting of ES cell clones with 5ʹ (upper panel) and 3ʹ probes (lower panel). Arrows indicate the target allele band. The asterisk indicates the clones used for generating chimeric mice. (C) Representative results of PCR genotyping of tail DNA of offspring obtained from heterozygous intercrosses. (D) Western blotting of eIF3e in the liver lysates of eIF3e +/+ (+/+) and eIF3e +/− (+/−) mice, which is representative of four animals of each genotype analyzed. GAPDH was used as a loading control. Columns represent mean values with SD. Asterisk indicates P < 0.05 when compared to eIF3e +/+. (E) Genotyping of mice generated from heterozygous intercrosses.
