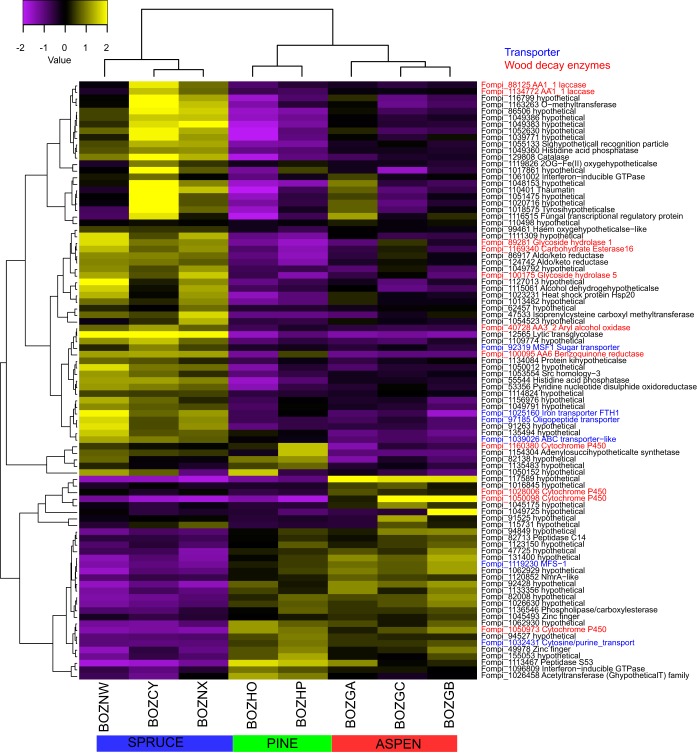
FIG 1.
Heatmap showing hierarchical clustering of 11 genes with predicted functions in wood decay with 4-fold change (FDR < 0.05) in pairwise-substrate comparisons at day 5. The scale above the map shows log2-based signals under the central row. Protein identifiers (IDs) and their enzyme names are indicated on the right side of the heatmap. Transporters are labeled in blue, and wood-decay enzymes are labeled in red. The bottom column designations refer to replicate libraries.