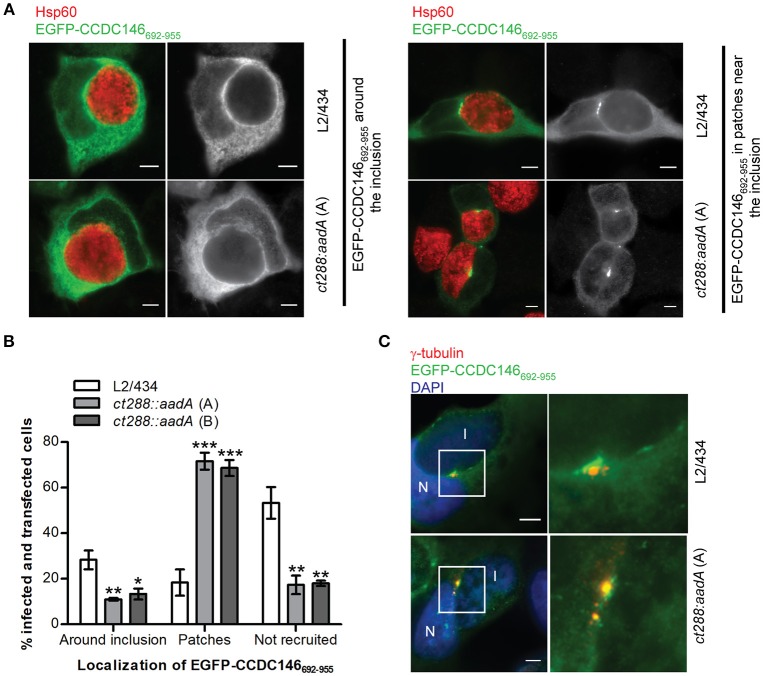
Figure 6.
Recruitment of EGFP-CCDC146652−955 to the periphery of the C. trachomatis inclusion is modulated by CT288. HeLa cells transfected with a plasmid encoding EGFP- CCDC146652−955 were infected for 24 h with C. trachomatis L2/434 or the ct288::aadA mutant (clones A and B; see Figure 5). The cells were fixed with methanol, immunolabelled with anti-GFP, anti-γ-tubulin, or anti-Hsp60 antibodies, and appropriate fluorophore-conjugated secondary antibodies, and/or stained with DAPI (as indicated), and analyzed by immunofluorescence microscopy. (A) Representative images of the localization of ectopically expressed EGFP-CCDC146652−955 surrounding the inclusion (left-side panels), or in patches near the inclusion (right-side panels), in Chlamydia-infected cells. (B) Enumeration of cells infected by either C. trachomatis L2/434 or the ct288::aadA mutant showing ectopically expressed EGFP-CCDC146652−955 surrounding the inclusion, in patches near the inclusion, or not recruited to the inclusion periphery. Data represents three independent experiments (at least 50 infected and transfected cells were counted per experiment). P-values were obtained by one-way ANOVA and Dunnett post-hoc analyses relative to cells infected by the L2/434 strain in each class (around inclusion, in patches, or not recruited). *P < 0.05; **P < 0.01; ***P < 0.001. (C) Co-localization between EGFP-CCDC146652−955 in patches near the inclusion and γ-tubulin, in cells infected by C. trachomatis L2/434 (less frequently observed; see B) or the ct288::aadA mutant (more frequently observed; see B). I, inclusion; N, host cell nucleus. Scale bars, 5 μm.