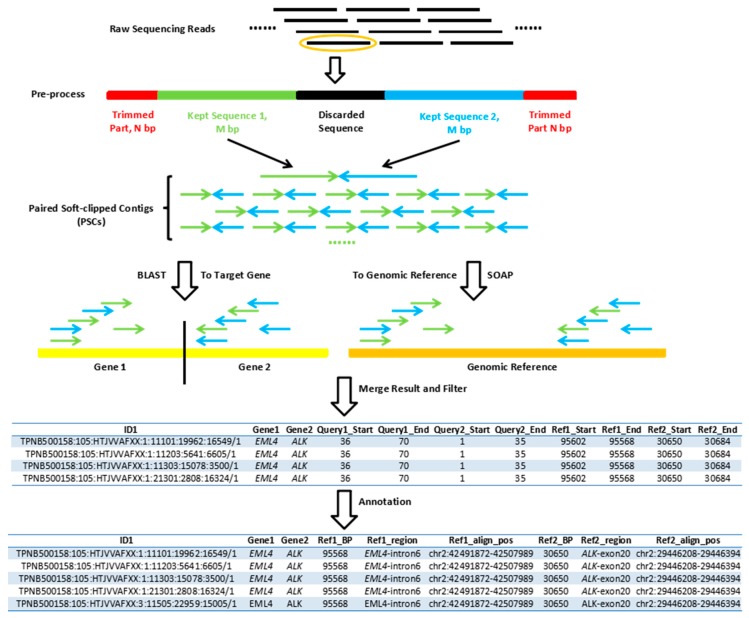
Figure 1.
The scheme shows how the single-end gene fusion (SEGF) works. Firstly, there is pre-processing of raw sequencing data, including trimming of the first and last N bp (red part) and merging the first and last M bp of the remaining sequence as paired soft-clipped contigs (PSCs) (green and blue part); the last remaining part (black part) was discarded, and not used in the following analysis. Basic local alignment search tool (BLAST) and Short Oligonucleotide Analysis Package (SOAP) were used to align PSCs into target gene references (yellow part) and genomic references (orange part) separately, keeping the result unique and fully mapped to reduce the influence of genomic repetitive regions. The mutual sequences of the two filtered results are considered as fusion sequences. If the number of mutual reads was larger than three, then the sample was considered as fusion positive, otherwise, fusion negative.