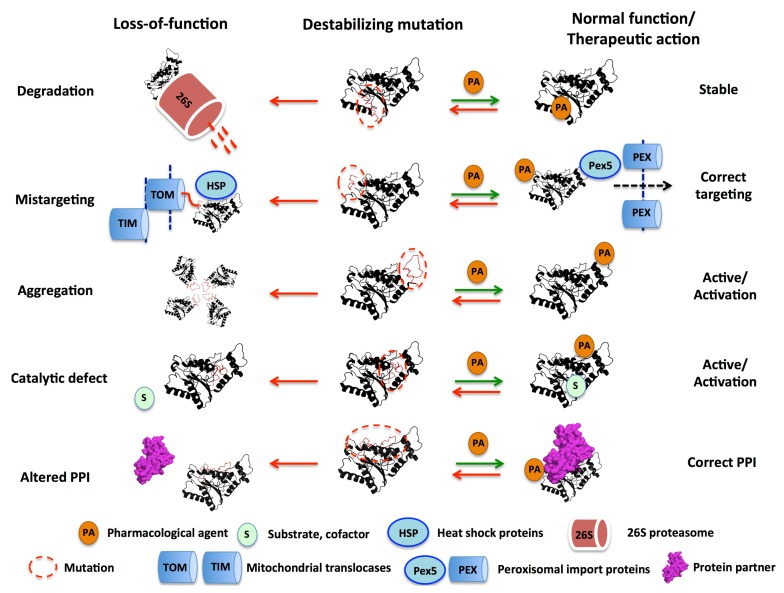
Figure 1.
Schematic representation of five of the main molecular mechanisms of loss-of-function (LOF) diseases associated with destabilizing mutations and relevant examples discussed in this review: (i) accelerated protein degradation by the 26S proteasome (e.g., the polymorphism P187S in NQO1; see Section 4.1.); (ii) intracellular mistargeting to mitochondria due to enhanced interaction with mitochondrial import machineries instead of normal targeting to peroxisomes upon interaction with Pex5p dependent pathway (e.g., variant G170R of AGT associated with PH1; see Section 4.2.); (iii) enhanced formation of inactive aggregates (e.g., variant I244T of AGT associated with PH1; see Section 4.2.); (iv) catalytic defect (e.g., the polymorphism P187S in NQO1 promotes the formation of the inactive apo-enzyme by altering FAD binding; see Section 4.1.); (v) alteration in protein:protein interactions (e.g., the polymorphism P187S reduces NQO1 steady-state levels thus destabilizing transcription factors such as p53 and p73α; see Section 4.1.). On the right side, the potential correction of local destabilizing effects by a pharmacological agent (e.g., a small ligand) is presented as promising approaches to rescue the function of disease-associated variants. AGT, Alanine:glyoxylate aminotransferase; PH1, primary hyperoxaluria type I; NQO1, NADP(H):quinone oxidoreductase 1; PPI, protein:protein interaction.