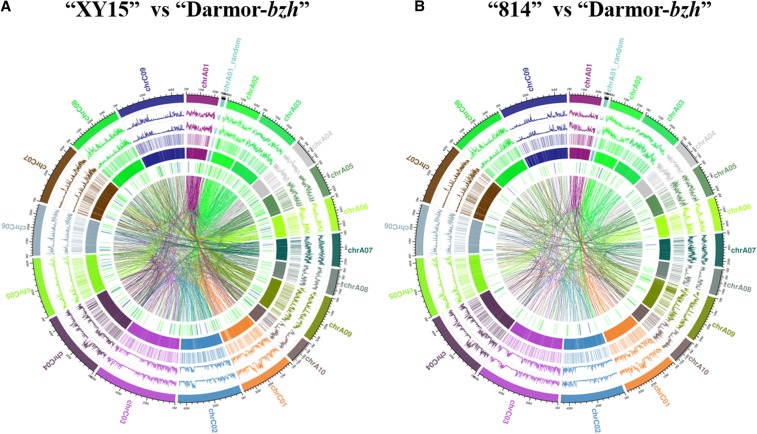
Figure 1.
Overview of the genetic diversity between the whole-genome sequenced rapeseed cultivars (N-efficient cv. “XY15” and N-inefficient cv. “814”) and the de novo sequenced reference genome of “Darmor-bzh”. Overview of the genome-wide genetic variants between “XY15” vs. “Darmor-bzh” (A) and “814” vs. “Darmor-bzh” (B), which are delineated by the Circos program. In the Circos figure, the variants are as follows outside-to-inside: chromosomes (i), single nucleotide polymorphisms (SNPs) (ii), insertions/deletions (InDels) (iii), copy number variation (CNV) duplications (iv), CNV deletions (v), structure variations (SV; red: insertions; green: deletions; blue: inversion) (vi), and intra-/inter-chromosomal translocation (vii).