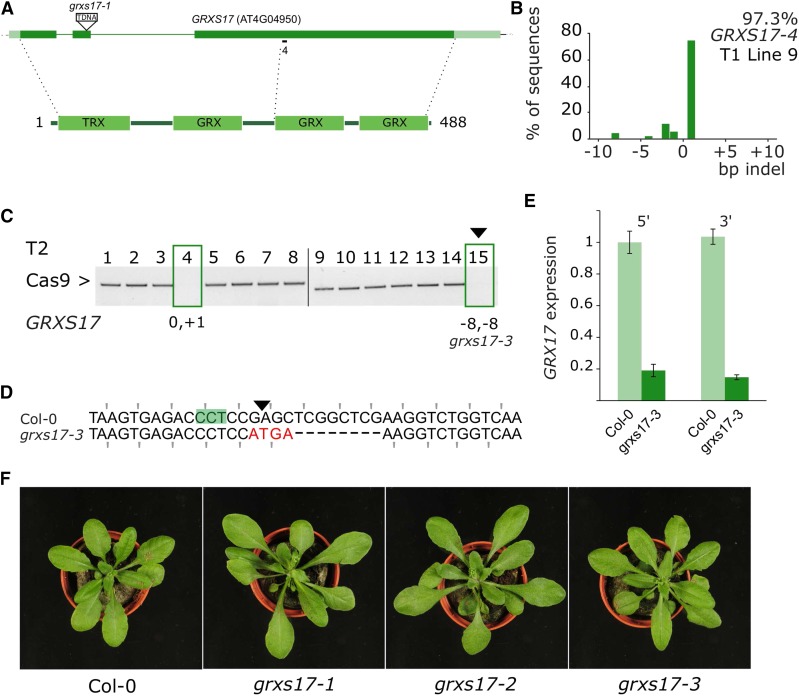
Figure 4.
Generation and analysis of the grxs17-3 allele. A, Gene and protein structure of GRXS17. The location of the grxs17-1 T-DNA and the sgRNA used in this study are indicated. Dark green boxes designate exons; light green boxes, UTRs; solid lines, introns. TRX, thioredoxin domain; GRX, glutaredoxin domain. B, TIDE analysis of T1 line 9. Genomic DNA was PCR amplified and sequenced. The indel spectrum is visualized with an estimated overall efficiency and the frequency of each indel. C, PCR amplification of the Cas9 transgene. Null-segregants are boxed and the continued plant marked with a triangle. TIDE estimated genotypes for GRXS17 are given for the null segregants. D, Sequence alignment of the targeted locus for Col-0 and grxs17-3 (Line 9, plant 15). PAM is highlighted, the Cas9 cut site indicated with a triangle. Mutated bases are in red, deleted bases replaced by an en dash. The reading frame is marked. E, GRXS17 gene expression analyzed by RT-qPCR. Expression relative to Col-0 is plotted using primers annealing both at the 5′ and the 3′ of the transcript and the mutation. F, rosette phenotypes of Col-0, the T-DNA insertion line grxs17-1, the antisense line grxs17-2 and the grxs17-3 CRISPR allele.