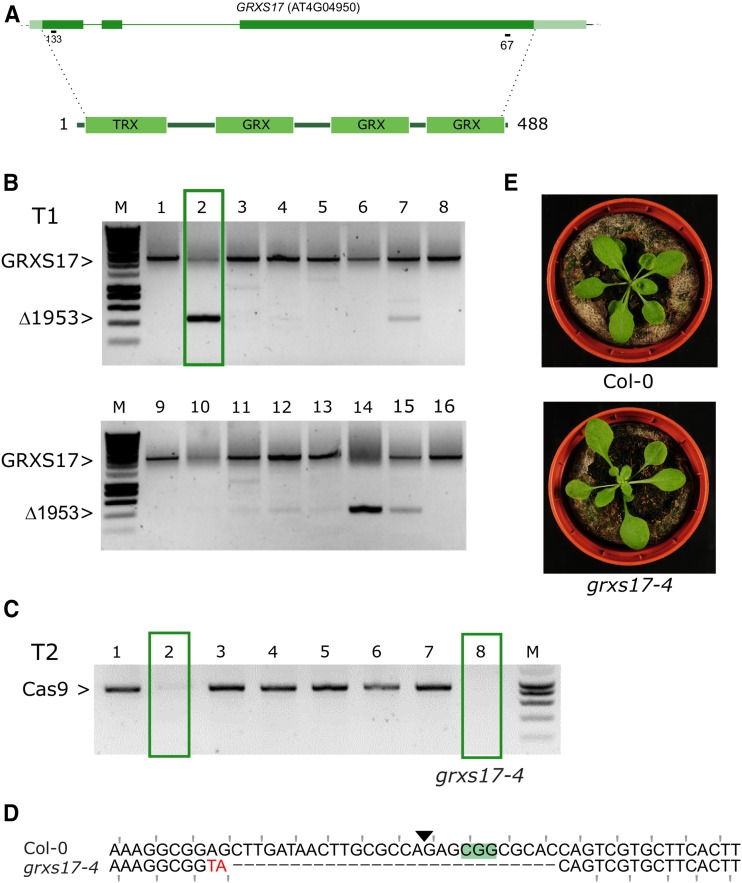
Figure 6.
A dual sgRNA approach for GRXS17. A, genomic structure of GRXS17 and location of the sgRNAs. Dark green boxes designate exons; light green boxes, UTRs; solid lines, introns. B, PCR analysis of T1 lines. Leaf genomic DNA of 16 chimeric T1 plants was PCR amplified. The expected size of the WT GRXS17 amplicon is indicated as well as the expected size of the deletion of 1953 bp between Cas9 cut sites. One T1 line having one T-DNA locus that was continued is highlighted with a green box. C, Cas9 PCR for 8 T2 CRISPR plants. Putative Cas9 null-segregants are indicated with green boxes. D, Sequence alignment of the sequence surrounding the 5′ sgRNA site for Col-0 and grxs17-4 (Line 2, plant 8). PAM is highlighted, the Cas9 cut site indicated with a triangle. Mutated bases are in red, deleted bases replaced by an en dash. The reading frame is marked. E, representative rosette phenotypes of WT Col-0 (top) and grxs17-4 (bottom).