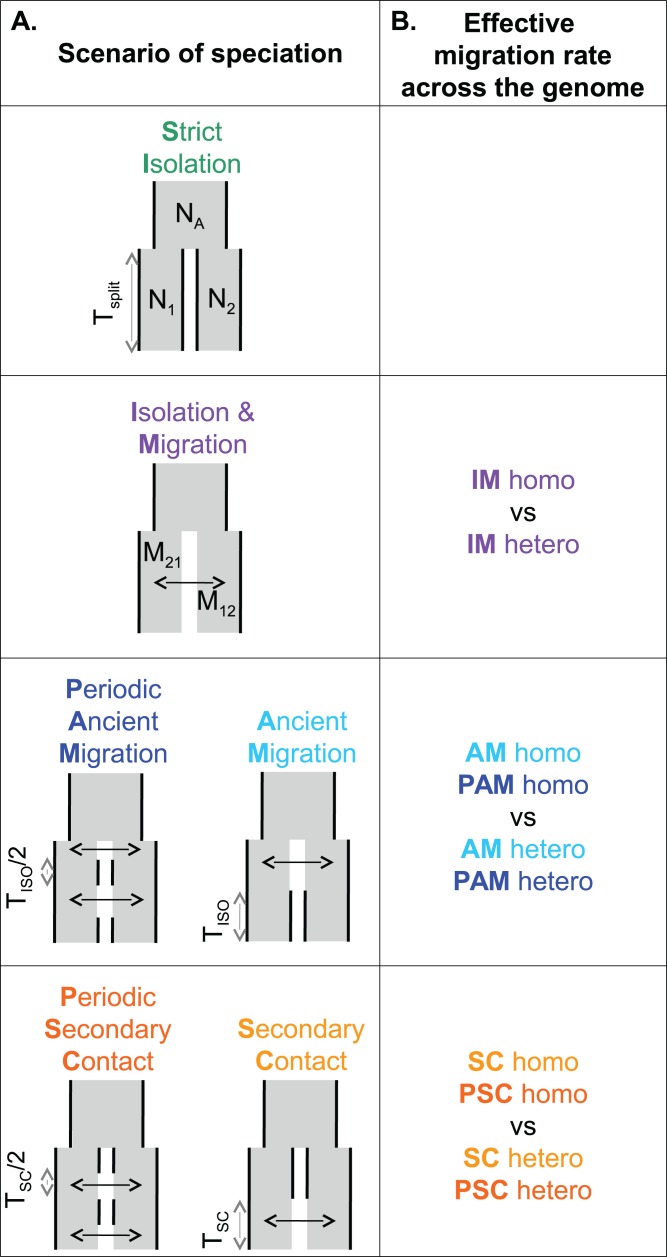
Figure 1. Models of speciation.
Six classes of scenarios with different temporal patterns of migration are compared (A); and for those including migration, two versions are depicted assuming either homogeneity (“homo”) or heterogeneity (“hetero”) of effective migration rate across the genome (B). All scenarios assume that an ancestral population of effective size NA split Tsplit generations ago into two populations of constant sizes N1 and N2. At the two extremes, divergence occurs in allopatry (SI, strict isolation) or under continuous migration (IM, isolation with migration). Through time, migration occurs at a constant rate M12 from population 1 to population 2 and M21 in the opposite direction. Ancient migration (AM) and periodic ancient migration (PAM) scenarios both assume that populations started diverging in the presence of gene flow. Then they experienced a single period of isolation, Tiso, in the AM model while intermittent gene flow occurred in the PAM model. In the secondary contact (SC) and periodic secondary contact (PSC) scenarios, populations diverged in the absence of gene flow followed by a single period of secondary contact, Tsc, in the SC model while intermittent gene flow occurred in the PSC model.