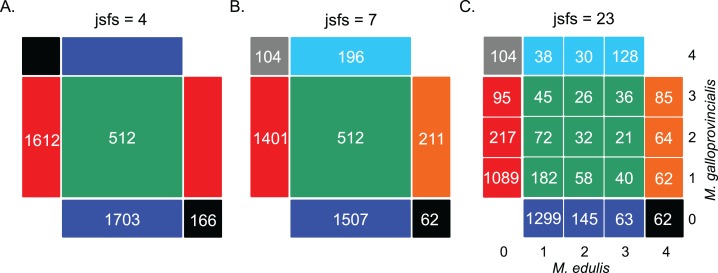
Figure 2. Decomposition of the unfolded joint site frequency spectrum for n = 2 individuals (i.e., four alleles) in each species.
The density of derived alleles in species 1 (M. edulis, x axis) and species 2 (M. galloprovincialis, y axis) is indicated by a number within each cell. Only sites showing two distinct alleles in the inter-specific alignment were considered, hence the cells {0; 0} and {4; 4} have been masked. The total number of polymorphic sites is 3,993 SNPs (“exome capture” data). (A) Decomposition of the jSFS into four classes of polymorphism without an outgroup sequence (i.e., the Wakeley–Hey classes): fixed differences (black), private polymorphisms in species 1 (blue) or species 2 (red) and shared polymorphisms (green). (B) Decomposition of the jSFS into seven classes of polymorphism by using the sequenced outgroup. Two alleles are differentially fixed between the two species: the derived allele can be fixed in species 1 (black) or in species 2 (gray). Exclusive polymorphism can be the result of a recent mutation specific to species 1 (blue) or species 2 (red); but it can also be the result of an ancestral mutation only fixed in species 2 (cyan) or in species 1 (orange). Shared polymorphisms are shown in green. (C) Decomposition of jSFS into 23 classes of polymorphism. Singletons and doubletons in each species were included as new classes. Note that in the case of n = 2, this is the full spectrum.