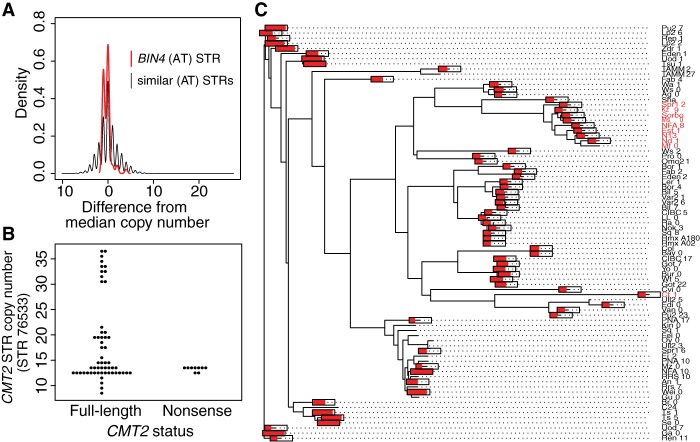
Figure 4.
Noncoding STRs showing non-neutral variation. (A) BIN4 intron STR is constrained relative to similar STRs. Allele frequency spectra are normalized by subtracting the median copy number (9 for the BIN4 STR). All pure STRs with TA/AT motifs and a median copy number between 7 and 12 are included in the “similar STRs” distribution. (B) Lack of association between near-expansion CMT2 STR alleles and previously described nonsense mutations. (C) Neighbor-joining tree of a 10-kb region of A. thaliana Chromosome 4 encompassing the CMT2 gene across 81 strains with available data, using Kimura's two-parameter distance model in APE (Paradis et al. 2004). Text labels in red indicate an adaptive nonsense mutation early in the first exon of CMT2, as noted previously (Shen et al. 2014). Red bars drawn on tips of the tree indicate the length of the CMT2 intronic STR (as a proportion of its maximum length, 36.5 units) in each of the 81 strains or tips. The bars are omitted for tips with missing STR data.