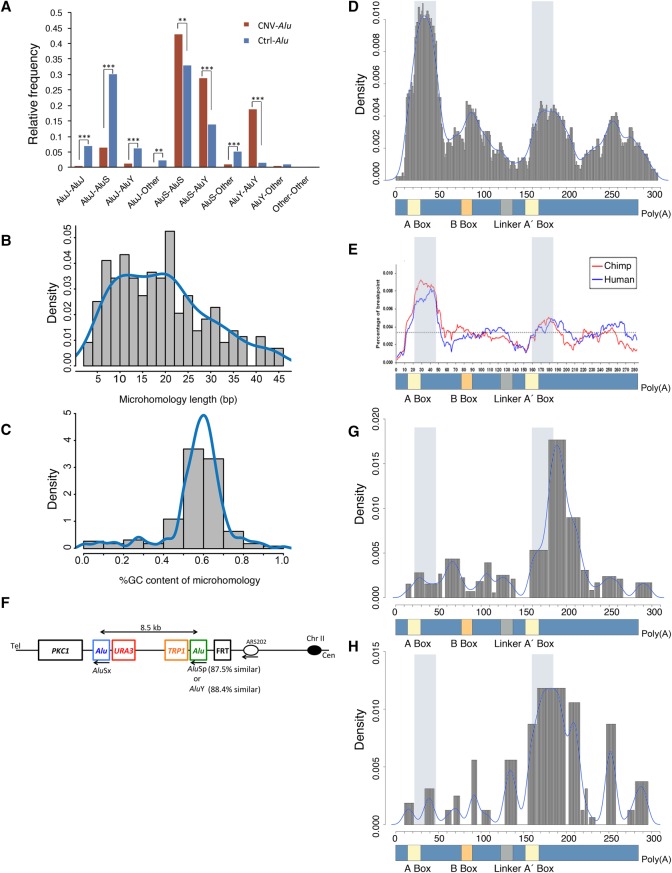
Figure 3.
Features of CNV-Alu pairs and microhomology preferences. (A) The relative frequency of Alu subfamilies is shown. For example, the AluS-AluY indicates CNVs mediated by Alus from family AluS and AluY respectively, and “Other” indicates monomeric Alus such as FRAMs. We compared the relative frequency of a given subfamily composition of CNV-Alu pairs (in maroon) with that of the expected relative frequency of observing a given subfamily pair (in blue) using the one-tailed binomial test. (**) P ≤ 0.01; (***) P ≤ 0.001. (B) The histogram describes the distribution of microhomology length at breakpoint junctions. (C) The histogram indicates the %GC content within the stretch of microhomology. (D) The figure depicts the collected 219 microhomologies from disease-related studies in human with respect to their relative position on an Alu consensus sequence (lower panel). The peak in the histogram indicates an enrichment of breakpoint junctions on the specific locus. The light blue shading shows a 26-bp core sequence detected by a previous compilation study of Alu-involved gene rearrangements (Rudiger et al. 1995). (E) Adapted from a comparative genomic study on chimpanzee and human reference genome (Han et al. 2007). The blue line describes 492 human-specific breakpoint junctions of Alu/Alu-mediated deletions, and the red line depicts 663 chimpanzee-specific events. The dashed horizontal line indicates the average percentage of breakpoints across the entire Alu element. (F) The schematic shows the construct utilized to detect template switches in yeast. Two human Alu pairs were inserted into Chr II separately with the same distal AluSx element. URA3 and TRP1 are the markers for selecting colonies with successful transformation. We induced a single-strand DNA break at the FRT site using a mutation of FLP recombinase. (G,H) The relative positions of microhomologies generated by mapping junctions from the yeast assay are depicted in relation to an Alu consensus sequence. (G) Data from 503 AAMR events observed in the first AluSx-AluSp strain. (H) Distribution of 114 events from the AluSx-AluY construct.