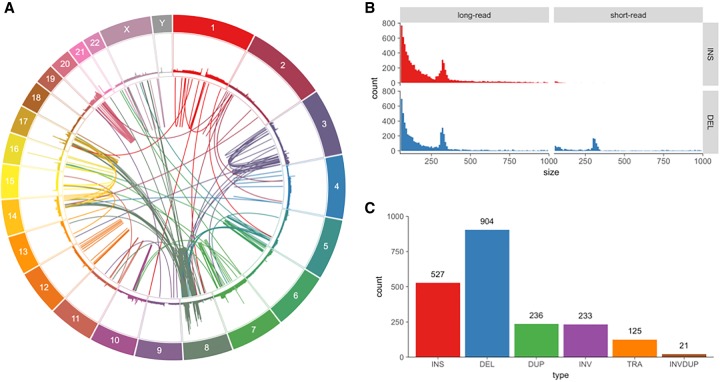
Figure 1.
Variants found in SK-BR-3 with PacBio long-read sequencing. (A) Circos (Krzywinski et al. 2009) plot showing long-range (larger than 10 kbp or inter-chromosomal) variants found by Sniffles from split-read alignments, with read coverage shown in the outer track. (B) Variant size histogram of deletions and insertions from size 50 bp up to 1 kbp found by long-read (Sniffles) and short-read (SURVIVOR 2-caller consensus) variant calling, showing similar size distributions for insertions and deletions from long reads but not for short reads, where insertions are greatly underrepresented. (C) Sniffles variant counts by type for variants above 1 kbp in size, including translocations and inverted duplications.