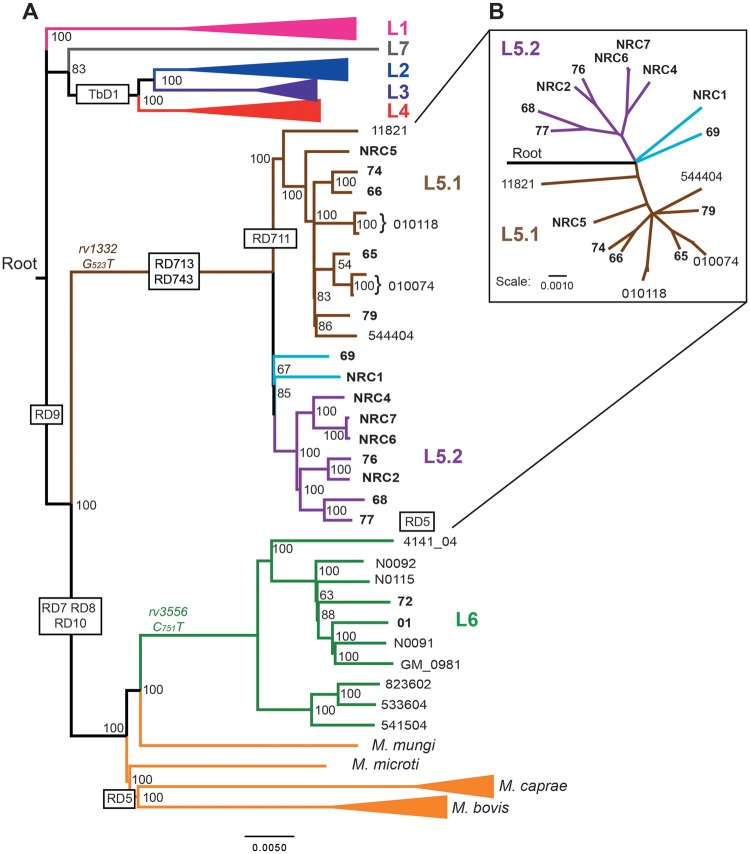
Fig. 1.
—Phylogenomic analysis of sequenced and previously published isolates reveals three sublineages within M. africanum L5. (A) Bootstrap consensus tree based on 32,510 variable positions inferred from 1,000 bootstrap replicates is taken to represent the evolutionary history of the isolates (Felsenstein 1989). Previously published genomes and associated accession numbers are listed in supplementary table 3, Supplementary Material online. The phylogenomic analysis was performed with RaxML and is based on variable positions identified with respect to M. tuberculosis H37Rv ( Stamatakis 2006, 2014). Lineages 1, 2, 3, and 4 as well as M. bovis and M. caprae strains are depicted as collapsed branches. A strain of the nonclonal, smooth tubercle bacilli (Blouin et al. 2014; Boritsch et al. 2014) known as Mycobacterium canettii, was used to establish the root of the tree, but is not depicted. Large sequence polymorphisms specific for the (sub)lineages are depicted within black boxes. SNPs specific for either L5 or L6 are depicted in brown or green, respectively. (B) Radial tree of L5 strains depicts the clear separation of L5.1 and L5.2 and the intermediate position of the orphan L5 isolates (light blue). Isolates sequenced in this study are in bold typeset. Newly proposed sublineages are color coded in the tree as: L5.1 (brown) and L5.2 (Lilac). Scale bars depict SNPs/1,000 base pairs.