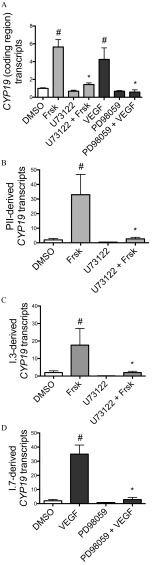
Figure 2.
Relative expression of (A) CYP19 coding region (nonpromoter-specific or total), and CYP19 transcripts derived from promoters (B) PII, (C) I.3, and (D) I.7 in Hs578t cells (fold DMSO control). Cells were exposed for 24 h to forskolin (Frsk) or VEGF, inducers of PII/I.3 or I.7 promoter-mediated CYP19 expression, in the presence or absence of selective inhibitors of the PLC (U73122; ) or MEK/MAPK 1/3 (PD98059; ) signaling pathways. Experiments were performed in triplicate with three different cell passages; per experiment, each treatment was tested in triplicate. *, . Statistically significant difference between Hs578t cells pretreated with U73122 compared with those treated with Frsk alone, or between Hs578t cells pretreated with PD98059 compared with those treated with VEGF alone (Student t-test). #, . Significantly different from DMSO control (Student t-test).