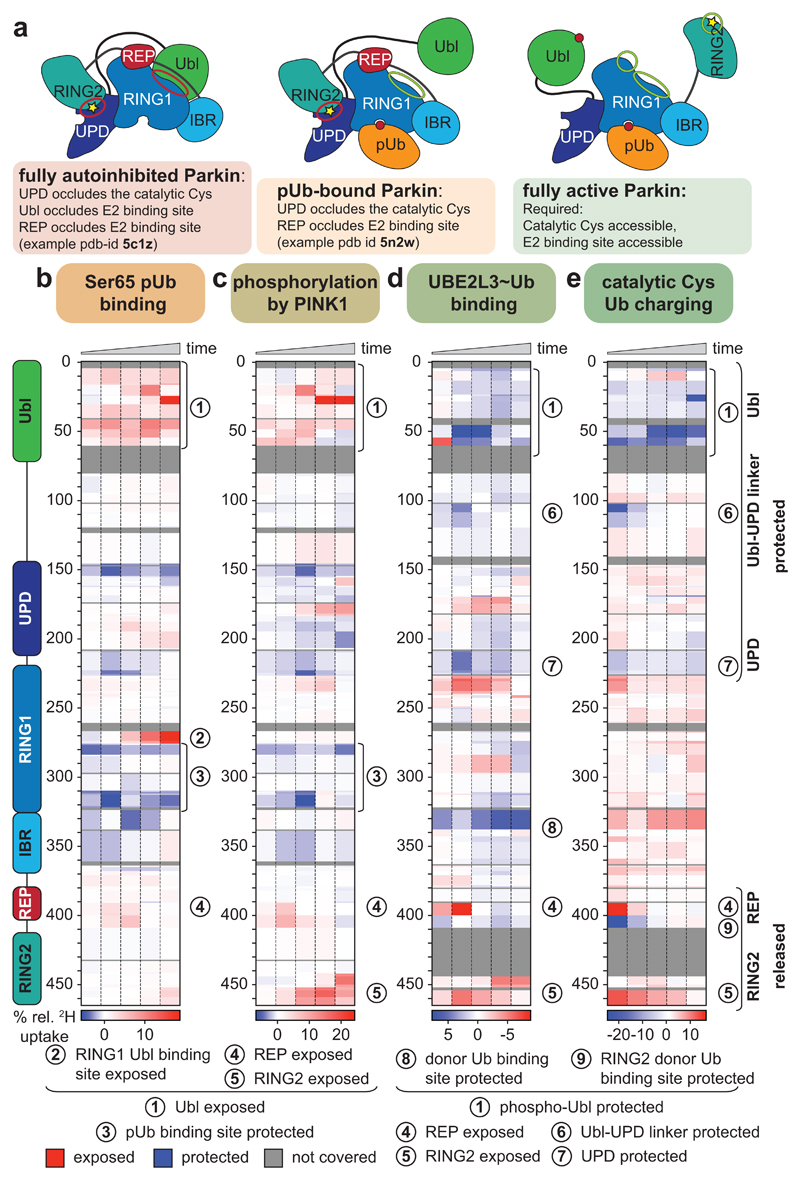
Figure 1. Domain rearrangements in Parkin, resolved by HDX-MS.
a, Cartoon of Parkin activation. Left, Parkin is autoinhibited by several mechanisms (red circles)7–9. Middle, phospho-ubiquitin binding to Parkin releases the Ubl domain, but most mechanisms of autoinhibition remain5,6. Right, after Ubl phosphorylation, Parkin is fully active (green circles), but a structure of active Parkin has not been reported. Also see Extended Data Fig. 1. b-e, HDX-MS difference map with the shortest peptides covering any given region, coloured from blue (more protected from exchange compared to previous state) to red (more accessible to solvent exchange), peptides for grey coloured regions could not be analysed (see Extended Data Fig. 3d). The five rows per sample indicate different time lengths for HD exchange (0.3 s, 3 s, 30 s, 300 s and 3000 s). All experiments were performed with human full-length Parkin, as technical triplicates. See Extended Data Fig. 2 and 3 for raw data and structural mapping, respectively. b, Difference between Parkin and Parkin bound to phospho-ubiquitin. c, Difference between Parkin:phospho-ubiquitin and phospho-Parkin:phospho-ubiquitin. d, Difference between phospho-parkin:phospho-ubiquitin, and phospho-Parkin:phospho-ubiquitin in complex with a non-dischargeable UBE2L3-Ub complex (see Online Methods). e, Difference between phospho-Parkin:phospho-ubiquitin and phospho-Parkin:phospho-ubiquitin charged with Ub-VS (see Online Methods).