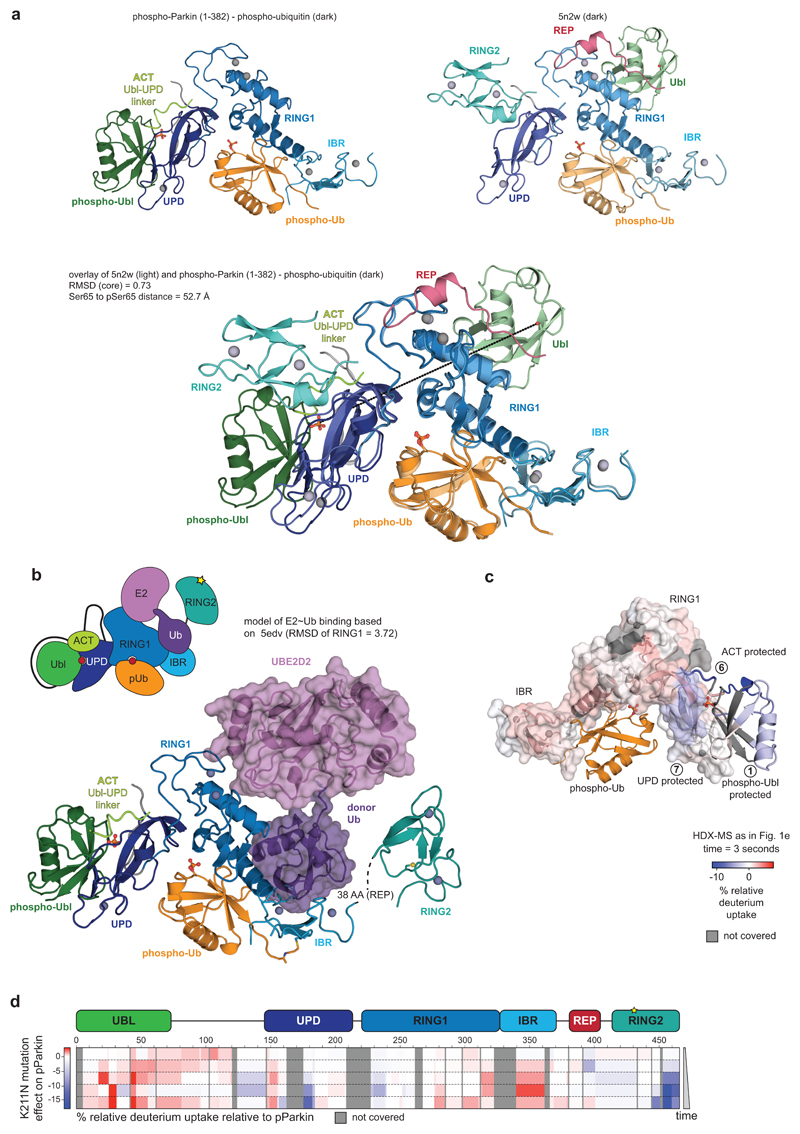
Extended Data Figure 7. The phospho-Ubl binding site on the UPD.
a, Side by side view of phospho-Parkin-pUb (left) and Parkin-pUb (pdb-id 5n2w, 6, right), and superposition of both (below). The green Ubl domain changes position by >50 Å. b, E2~Ub from the structure of HOIP RBR domain in complex with UBE2D2~Ub29, was modelled onto phospho-Parkin-pUb, by superposition of the RING1 domains of each complex. The E2-conjugated ubiquitin molecule in the ‘open’ conformation binds to the previously recognised cryptic ubiquitin binding interface on RING1/IBR 6. The contact points correlate with HDX-MS data (Fig. 1d, Extended Data Fig. 2, 3). c, HDX-MS data from Fig. 1e was plotted onto the phospho-Parkin-pUb structure with identical colouring (blue, more protected from solvent exchange; red less protected from solvent exchange; grey, not covered in all of the compared states, compare Fig. 1). Protected regions on UPD match the observed phospho-Ubl interface. d, HDX-MS experiments comparing a Parkin mutant with a mutation in the phospho-acceptor binding site on the UPD, (phospho-Parkin K211N:phospho-Ub) compared with phospho-Parkin:phospho-Ub, coloured as in c. The mutant is unable to protect the Ubl, and to release RING2 and REP. Experiments were done as technical triplicate.