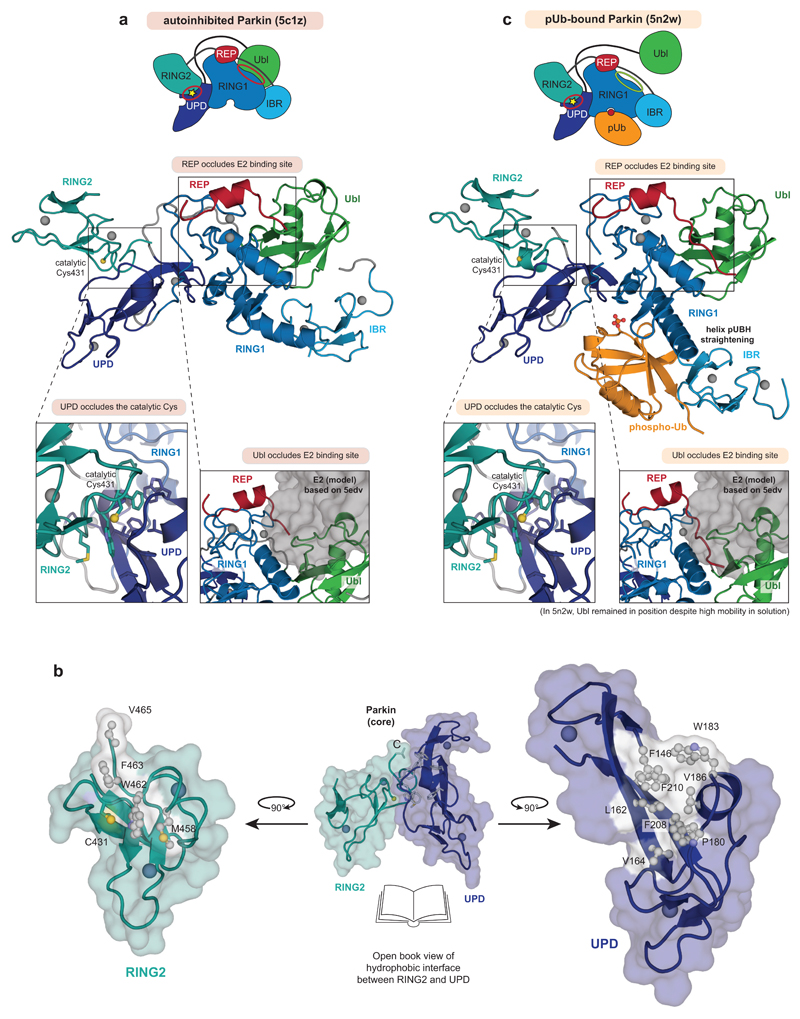
Extended Data Figure 1. Mechanisms of Parkin autoinhibition.
a, Structure of autoinhibited, full-length human Parkin (pdb-id 5c1z, 19) shown schematically (top, as in Fig. 1a) and in cartoon representation in the same colours. Two insets show the UPD-RING2 interface (with Cys431 shows in ball-and-stick representation), and the blocked E2 binding site (with the E2 position, modelled according to 5edv 29 shown as grey surface). Zn ions are shown as grey spheres. b, An ‘open-book’ view on the UPD-RING2 interface, with hydrophobic residues coloured white on each surface. c, Structure of phospho-ubiquitin bound to full-length Parkin (pdb-id 5n2w, 6) as in a. Phospho-ubiquitin binding leads to helix straightening, and IBR domain repositioning, which releases the Ubl domain for phosphorylation5,6. In the shown structures of unphosphorylated Parkin, the Ubl and REP (red) inhibit E2 binding, and the RING2-UPD interface is intact, with Cys431 being inaccessible. The Ubl-UPD linker was removed from both crystallised constructs6,22.