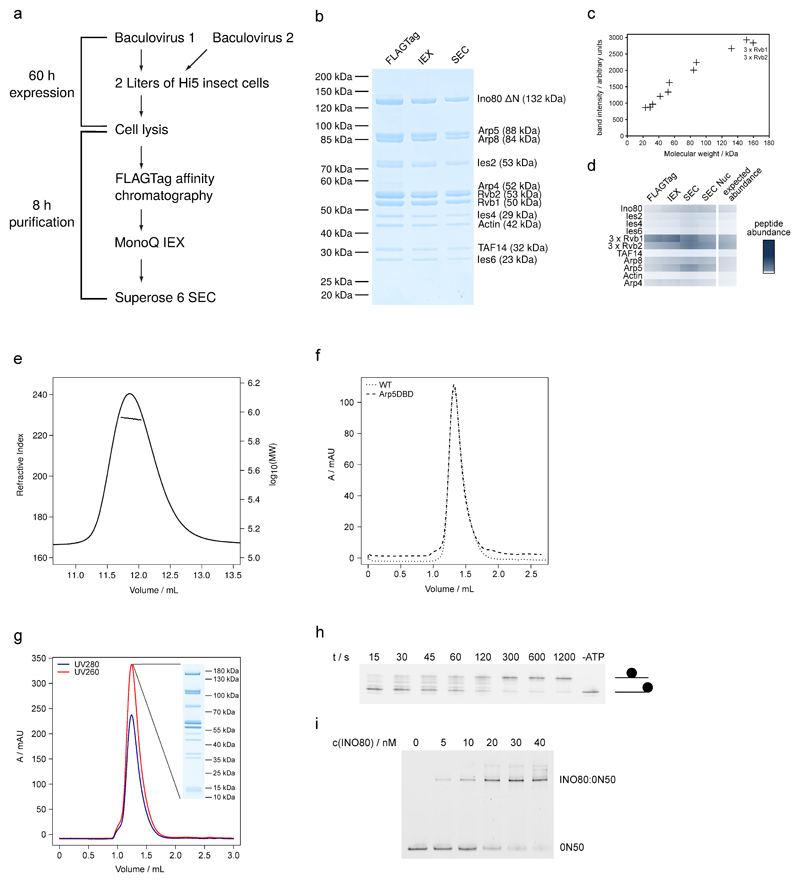
Extended Data Figure 1. Purification of apo-INO80, INO80:0N50 and sliding activity of INO80.
(a) Scheme of expression and purification of INO80.
(b) Documentation of INO80 purification by SDS PAGE (stained with simply blue). Protein identity was confirmed by mass spectrometry (data not shown).
(c) Quantification of band intensity from SDS PAGE (SEC sample) plotted against the molecular weight show stoichiometric presence of all subunits.
(d) Label free semi-quantitative mass spectrometry analysis of INO80core complexes after individual purification steps.
(e) RALS measurement of apo-INO80. Measured refractive index and calculated logarithmical molecular weight are plotted against the elution volume. The measurement yields a molecular weight of 880 kDa, confirming the integrity and correct stoichiometry of the purified complex.
(f) Comparison of the SEC elution profile of apo-INO80 and the Arp5DBD mutant on a Superose 6 3.2/300.
(g) Purification of the INO80:nucleosome complex. SEC elution profile of a Superose 6 3.2/300 is shown together with an analysis of the main peak fraction by SDS PAGE.
(h) Sliding of end-positioned 0N80 mononucleosomes by INO80. Native PAGE analysis of Fluorescein-labeled nucleosome is shown.
(i) Interaction of INO80 and mononucleosome monitored by electrophoretic-mobility shift assay.