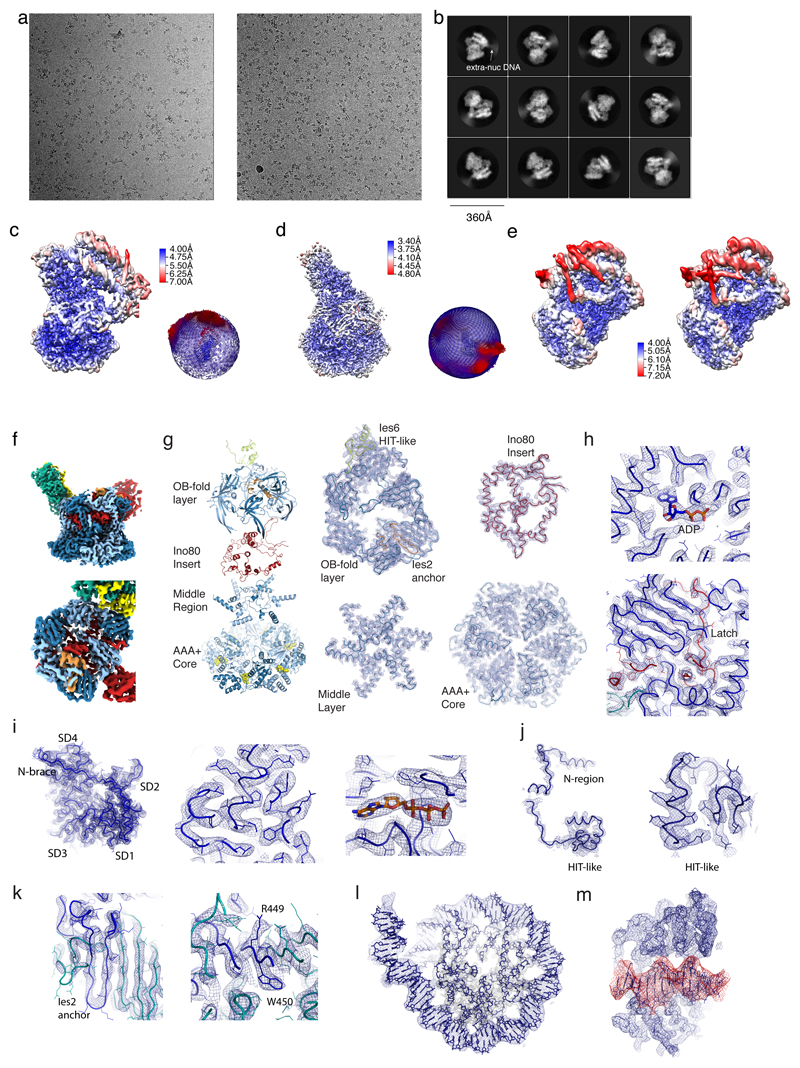
Extended Data Figure 3. Cryo EM data quality.
(a) Two representative micrographs of the set that was used to determine the structure of the INO80core:NCP complex.
(b) Typical 2D class averages of the INO80core:NCP complex. Note that dynamic extra-nucleosomal DNA (denoted as extra nuc DNA) visibly protrudes from the well-ordered core complex.
(c)-(e) The final 4.3Å (c, overall), 3.7Å (d, Rvb1/2-Arp5 mask) and 4.6Å and 4.7 Å (e, grappler conformations B and A) maps were analyzed by using ResMap48. Local resolution estimates are shown as a color-coded surface representation along with representations of angular distributions of particles contributing to the 4.3Å and 3.7Å maps.
(f)-(m) Representative examples of cryoEM map areas used for model building.
(f) The 3.7Å map using the color codes of Fig. 1c show the definition of Rvb1/2-client interactions.
(g) “Explosion” figure of the Rvb1/2 layers, along with corresponding regions of the 3.7Å map.
(h) Top: details showing a representative ATP/ADP binding site of Rvb1/2, with highlighted ADP, and showing the “latch” of the Ino80insert (red).
(i) Map at Arp5core, showing the N-terminal brace (left), with representative details of the actin core (middle) and the ATP binding site (right).
(j) Overview showing Ies6 (left) and details of its HIT-like domain (right).
(k) Map at the Ies2-Rvb1/2 interaction (left) with details showing an anchoring tryptophane.
(l) Map area at the NCP
(m) Map area at the Ino80 motor domain bound to SHL-6 (red).