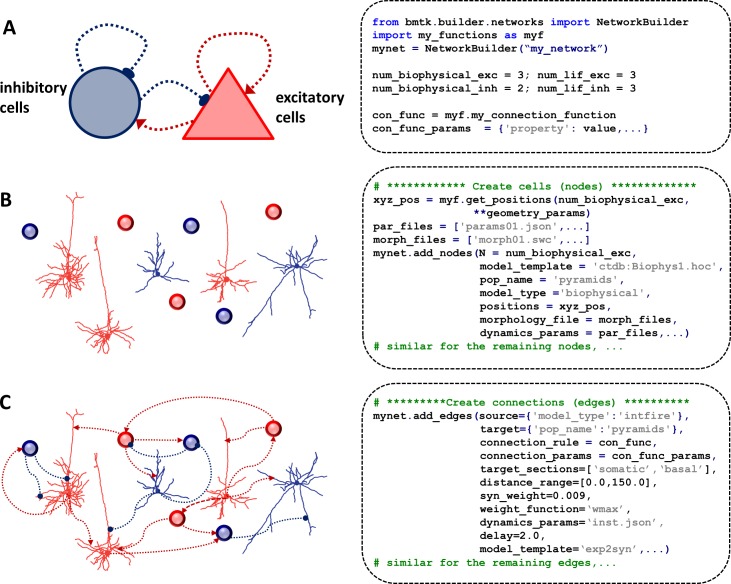
Fig 3. Building networks.
(A) High-level specification of a simple example network (left) and corresponding builder API commands (right). The model is composed of two cell types: inhibitory (blue) and excitatory (red), which exchange connections both across and between the cell types. The API commands define the number of cells of each type to be created, connectivity rule (con_func) to use and associated parameters (con_func_params) as well as additional edge parameters (edge_type_params). (B) Illustration of creating cells (left) where each cell type may include both biophysical (morphological reconstruction) and LIF models (circles). The corresponding API commands for adding nodes for the biophysically detailed subset of excitatory populations are illustrated on the right. Here we specify the number of nodes to be created (N), a type of a model (model_type), the dynamical cell models (model_template) and the corresponding model parameters (dynamics_params), morphologies (morphology_file), and positions of cell somata (positions) that were computed with a user-defined function. (C) Illustration of connecting the cells into a network (left) and the corresponding API commands for adding a particular subset of connections (right). Here, the cells satisfying the query for both the source and target nodes will be connected using a function (connection_rule) with parameters (connection_params). The additional edge_type attributes are shared across the added edges and include the synaptic strength (syn_weight), function modulating synaptic strength (weight_function), dynamical synaptic model (model_template) and corresponding parameters (dynamics_params), a conduction delay (delay), as well as the locations where synapses could be placed on a cell (target_sections, distance_range).