Abstract
Acute myocardial infarction induces ventricular remodeling, which is implicated in dilated heart and heart failure. The pathogenical mechanism of myocardium remodeling remains to be elucidated. The aim of the present study was to identify key genes and networks for myocardium remodeling following ischemia-reperfusion (IR). First, the mRNA expression data from the National Center for Biotechnology Information database were downloaded to identify differences in mRNA expression of the IR heart at days 2 and 7. Then, weighted gene co-expression network analysis, hierarchical clustering, protein-protein interaction (PPI) network, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were used to identify key genes and networks for the heart remodeling process following IR. A total of 3,321 differentially expressed genes were identified during the heart remodeling process. A total of 6 modules were identified through gene co-expression network analysis. GO and KEGG analysis results suggested that each module represented a different biological function and was associated with different pathways. Finally, hub genes of each module were identified by PPI network construction. The present study revealed that heart remodeling following IR is a complicated process, involving extracellular matrix organization, neural development, apoptosis and energy metabolism. The dysregulated genes, including SRC proto-oncogene, non-receptor tyrosine kinase, discs large MAGUK scaffold protein 1, ATP citrate lyase, RAN, member RAS oncogene family, tumor protein p53, and polo like kinase 2, may be essential for heart remodeling following IR and may be used as potential targets for the inhibition of heart remodeling following acute myocardial infarction.
Keywords: weighted gene co-expression network analysis, ischemia-reperfusion, network, hub gene
Introduction
Although the medical approaches have developed rapidly, acute myocardial infarction (AMI) remains a principal cause of mortality worldwide (1). AMI induces a complex pathological processes including the death of cardiomyocytes, inflammation, fibrosis and hypertrophic growth of cardiomyocytes, which may progress to heart failure (2). Currently, the most effective therapies for AMI are vascular reflow to the ischemic region by thrombolysis, percutaneous coronary intervention and coronary artery bypass surgery (3).
As the ischemic area remains hypoxic, the heart continues to lose cardiomyocytes, which decreases its contractile function. The condition can be modeled by the permanent ligation of coronary artery in rodent animals. When the myocardium is reperfused for the reoxygenation of ischemic myocardium, the reoxygenation can induce reperfusion injury through cell injury pathways, including free radical production, mitochondrial injury, activation of pro-apoptotic pathway and calcium dysregulation (4). Reoxygenation also induces cardiac remodeling by the transdifferentiation of cardiac fibroblasts into myofibroblasts (4–6). These side effects of reperfusion may increase the risk of mortality (7,8). Therefore, a comprehensive understanding of myocardial biology in ischemia-reperfusion (IR) is important.
Weighted gene co-expression network analysis (WGCNA) has been successfully used in systems biology studies to investigate the intrinsic organization of transcriptomes (9). This approach provides an effective method to examine the gene expression patterns, calculate the adjacency of genes, construct gene networks and assess the importance of genes within the network (10). It has been applied for construction of gene co-expression network modules, the identification of hub genes and identifying the connection of diagnostic genes and diseases (10–12). However, WGCNA has not been applied to identify the network and hub genes associated with IR myocardium from days 2 to 7, to the best of the authors' knowledge.
In the present study, the microarray data on IR remodeling were downloaded from National Center for Biotechnology Information (NCBI) (13) and differentially expressed genes (DEGs) identified by pairwise comparison. Subsequently, WGCNA was performed to investigate the expression modules and key genes for each module. The functional annotation of each module revealed the biological associations with dynamic features of myocardium from days 2 to 7.
Materials and methods
Analysis of differentially expressed genes
The microarray data of rat IR were downloaded from the NCBI Gene Expression Omnibus database (accession number: GSE4105) and normalized by MAS5 (13). All the samples were divided into 4 groups, including day 2 (2d)-sham, 2d-IR, day 7 (7d)-sham and 7d-IR. DEGs were analyzed between two subsequent groups. DEGs between any two consecutive stages were determined by the limma 3.34.9 package in R (version 3.1.1) (14). The threshold of the significance of DEG was set as fold change 31.5 and P<0.05. A total of 3,321 DEGs were obtained with the cutoff set as P<0.05.
Construction of gene co-expression networks
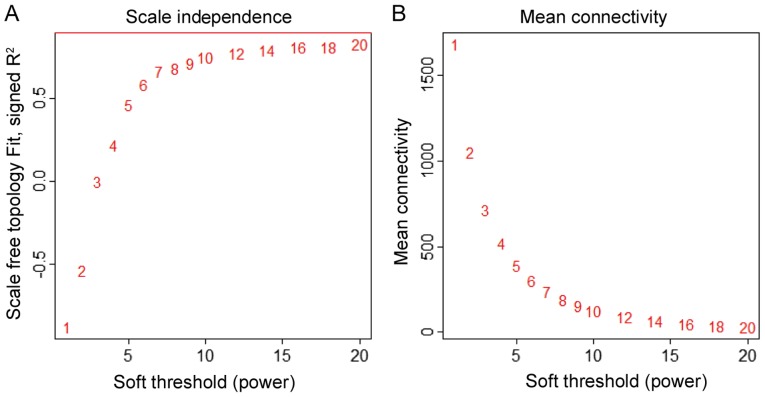
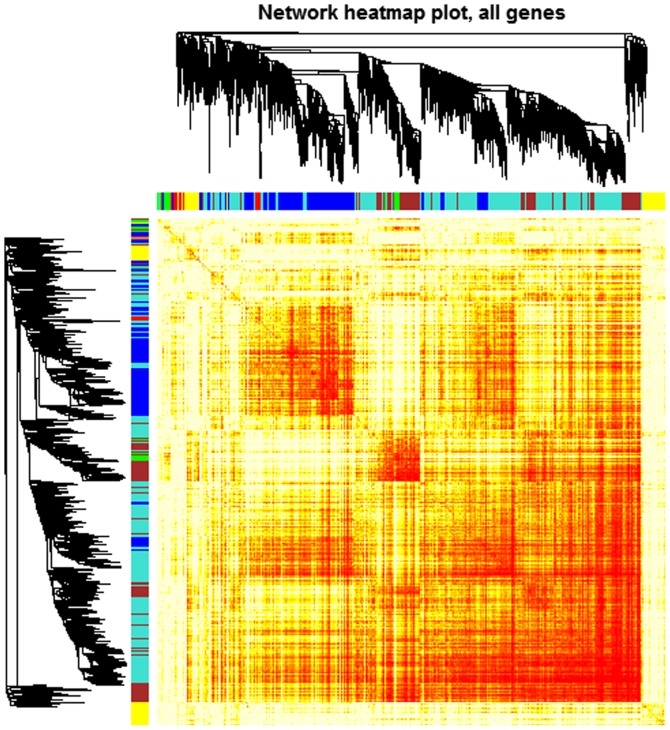
To study the functional organization of the rat IR heart, DEGs between any two consecutive stages were selected and weighted co-expression network analysis performed by using the WGCNA R package (15). First, missing values and outlier samples were checked by an unsupervised hierarchical clustering analysis. Then, a weighted adjacency matrix was created and the soft threshold power (β) set at 10 to analyze scale-free topology. The power β determines the co-expression similarity for the adjacency matrix. The power 10 was chosen, for which the scale-free topology fitting index R2 and mean connectivity may reach a plateau (Fig. 1). It suggested that β=10 was the minimum value for the scale-free topology criterion. Modules were identified by the following parameters: ‘power =10, minModuleSize =30, mergeCutHeight =0.25’. A total of 6 modules containing all the DEGs were identified. Genes of each module are demonstrated in the topological overlap heatmap (Fig. 2).
Figure 1.
Different soft-thresholding powers (b) were analyzed for (A) the topology scale-free fitting and (B) mean connectivity as a function of the power evaluated from ischemia reperfusion myocardium data.
Figure 2.
Heatmap of the gene coexpression network. The heatmap descriped adjacencies among genes in the analysis. A single gene correspond to each row and column of the heatmap. Low adjacencies are indicated by yellow colour and higher adjacencies are indicated by progressively red colour. The genes in the analysis were clustered in modules labeled by a color code (blue, yellow, brown, green and turquoise), which are shown by squares under gene dendrogram along the diagonal.
Modules were labeled using different colors and shown as a hierarchical clustering dendrogram. The weighted network was visualized using a heatmap plot. Gene expression patterns of DEGs in each module were analyzed by hierarchical clustering analysis.
Gene Ontology (GO) and pathway analysis
To identify the biological process and signaling pathways in each module, GO (16) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (17) pathways were enriched by David online tools (david.ncifcrf.gov) (18,19). The threshold of significant enrichment was set as P<0.05.
Protein-protein interaction (PPI) network analysis
To determine the interaction between DEGs in each module, they were mapped to Search Tool for the Retrieval of Interacting Genes (STRING; version 10.5), an online tool for evaluating protein-protein interaction (20). Interactions with a combined score >0.4 were considered significant. The protein-protein networks were constructed by Cytoscape software (version 3.2.0; http://www.cytoscape.org/) (21). Modules of protein-protein interaction were identified by Molecular Complex Detection (MCODE; version 1.5) (22) with the following cutoff: Nodes of cluster >4, Score of MCODE >3.
Results
Functional annotation of DEGs in each module
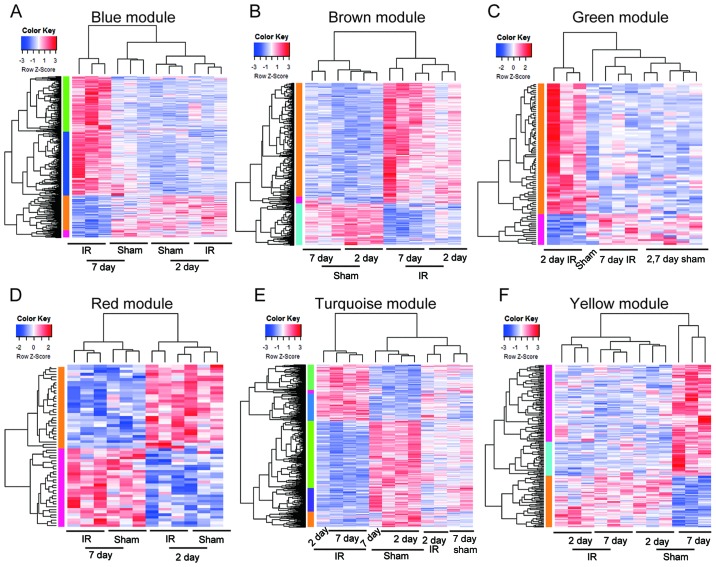
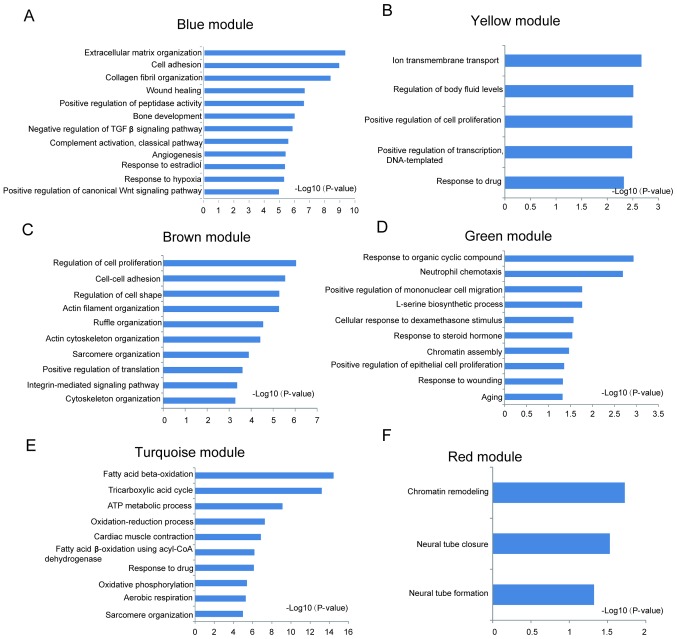
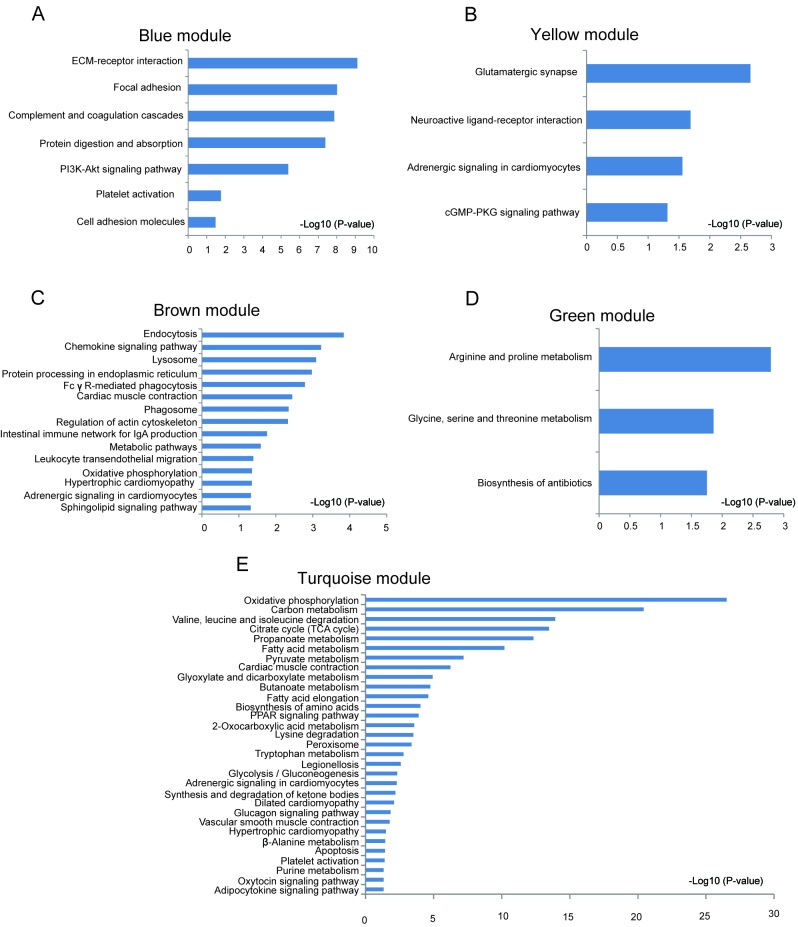
Significantly enriched biological process and KEGG pathways in each module are shown in Fig. 3. Different modules represented distinct gene expression patterns following IR. The blue module denoted the DEGs enriched between 7d-IR and other groups (Fig. 3A). ‘Extracellular matrix organization’, ‘wound healing’, ‘angiogenesis’, ‘canonical Wnt signaling’ and ‘PI3K-AKT signaling’ were significantly enriched in this module (Figs. 4A and 5A). In the brown module, DEGs were enriched between IR and sham group (Fig. 3B). ‘Regulation of cell proliferation’, ‘regulation of cell shape’, ‘intergrin mediated signaling pathway’, ‘endocytosis’, ‘Fc gamma R-mediated phagocytosis’, ‘cardiac contraction’, ‘hypertrophic cardiomyopathy’, ‘adrenergic signaling’ and ‘sphingolipid signaling pathway’ were significantly enriched in this module (Figs. 4C and 5C). In the green module, DEGs were enriched between 2d-IR and other groups (Fig. 3C). ‘Response to organic cyclic compound’, ‘epithelial cell proliferation’, ‘response to wounding’, and ‘amino acid metabolism’ were enriched significantly in this module (Figs. 4D and 5D). In the red module, DEGs were enriched between days 2 and 7 (Fig. 3D). ‘Chromatin remodeling’, ‘neural tube closure’, and ‘neural tube formation’ were enriched significantly in this module (Fig. 4F). In the turquoise module, the dynamic pattern of DEGs is shown in Fig. 3E. ‘Fatty acid β-oxidation’, ‘tricarboxylic acid cycle’, ‘ATP metabolic process’ and ‘oxidative phosphorylation’ were enriched significantly in this module (Figs. 4E and 5E). In the yellow module, DEGs were enriched between sham-7 day and other groups (Fig. 3F). ‘Ion transmembrane transport’, ‘regulation of body fluid levels’, ‘adrenergic signaling’ and ‘cGMP-PKG signaling’ pathways were significantly enriched (Figs. 4B and 5B).
Figure 3.
Hierarchical clustering analysis of all differentially expressed genes (DEGs) in each module. Blue color represents downregulation of genes. Red indicates upregulation of genes. Different gene expression patterns following IR were shown as different color modules. (A) The blue module enriched the DEGs between 7d-IR and other groups. (B) The brown module enriched DEGs between IR and sham group. (C) The green module enriched DEGs between 2d-IR and other groups. (D) The red module enriched DEGs between days 2 and 7. (E) The turquoise module enriched the dynamic expression of DEGs. (F) The yellow module enriched DEGs between sham-7 day and other groups. IR, ischemia reperfusion.
Figure 4.
Gene Ontology enrichment analysis of all differentially expressed genes (DEGs) in each module. DEGs in each module were subjected into biological process enrichment. Top 10 significantly enriched biological process were shown here (P<0.05) for (A) blue, (B) yellow, (C) brown, (D) green, (E) turquoise, and (F) red modules.
Figure 5.
Pathway enrichment of all DEGs in each module. DEGs in each module were performed KEGG enrichment. Significantly enriched KEGGs (P<0.05) are shown here for (A) blue, (B) yellow, (C) brown, (D) green and (E) turquoise modules. DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Identification of the key genes in each module
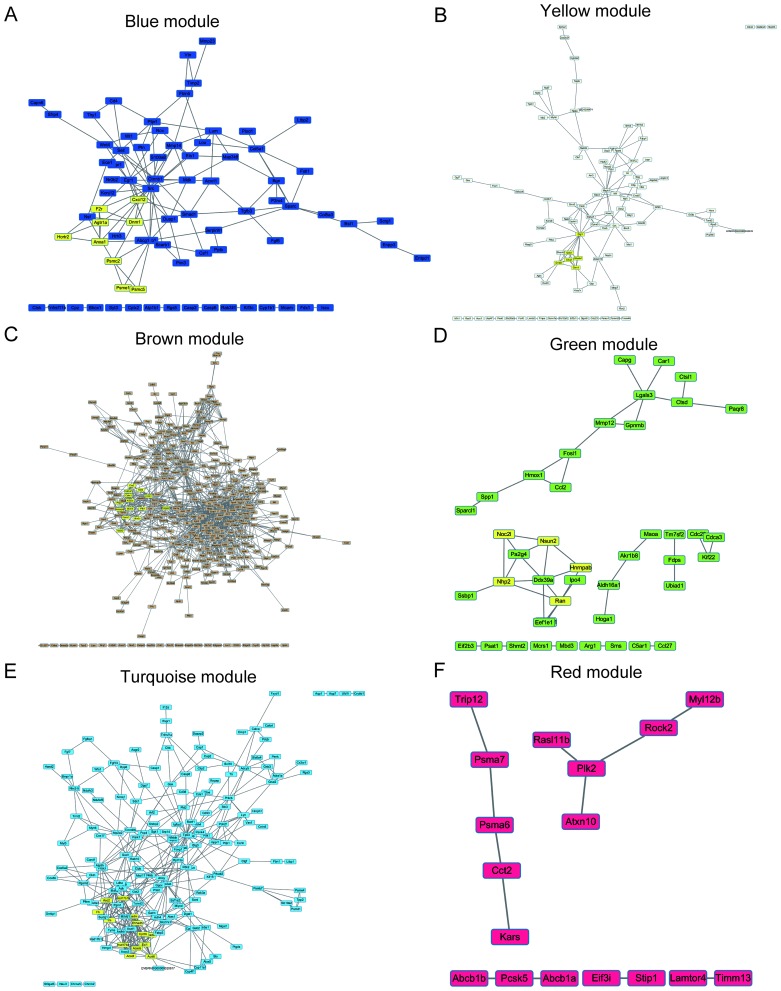
To elucidate the interaction between the hub genes in each module, PPI networks of hub genes were constructed using Cytoscape software. To explore core modules of protein-protein interactions, PPI modules were identified by MCODE.
The protein interacting with maximum number of DEGs was defined as the key gene in each module. As shown in Fig. 6, the key genes of each module are SRC proto-oncogene, non-receptor tyrosine kinase (Scr) for the blue module, discs large MAGUK scaffold protein 1 (Dlg1) for the yellow module, ATP citrate lyase (Acly) for the brown module, RAN, member RAS oncogene family (Ran) for the green module, tumor protein p53 (TP53) for the turquoise module and polo like kinase 2 (Plk2) for the red module. The most significant cluster of each network was highlighted for yellow color in each module.
Figure 6.
Protein-protein interaction network of the differentially expressed genes in each module. The nodes indicate proteins. The edges indicate interactions between proteins. Highlighted yellow nodes represent molecular complexes identified by molecular complex detection algorithm of cytoscape software. Core protein-protein interaction networks were shown here for (A) blue, (B) yellow, (C) brown, (D) green, (E) turquoise, and (F) red modules.
Discussion
The aim of the present study was to identify the genes and networks associated with the heart remodeling process following IR. Distinct modules, biological process, KEGG pathways and co-expression networks during heart remodeling were identified and the results of the present study provided insights into the regulation of heart remodeling following IR.
A total of 6 modules were identified by constructing co-expression networks. Hierarchical clustering analysis revealed that each module represented a different dynamic gene expression pattern and a different biological function. The blue module represented the specific physiological features of the heart following 7 days' IR, associated with ‘canonical Wnt signaling’, ‘TGFβ receptor signaling’ and ‘PI3K-AKT signaling pathway’. It has been reported that canonical Wnt signaling, TGFβ and PI3K-AKT signaling pathways are involved in regulation of cardiac fibroblasts growth and extracellular matrix (ECM) remodeling (23–26).
The brown module represented the common physiological features of the heart following IR, which are associated with phagocytosis. The green module represented the specific physiology of the heart after 2 days' IR, associated with amino acid metabolism. Apoptosis-associated genes were significantly upregulated in the day 2 IR heart, including heme oxygenase 1, FOS like 1, AP-1 transcription factor subunit and eukaryotic translation elongation factor 1 epsilon 1. The red module represented the physiological state of the heart between days 2 and 7, which is associated with neural development. The turquoise module represented the gradual process of heart remodeling which is closely associated with fatty acid metabolism. It has been documented that during heart remodeling and heart failure, the energy metabolism shifts from the fatty acid metabolism to glucose metabolism (27). In this module, fatty acid metabolism genes including acyl-CoA oxidase 3, pristanoyl, carnitine palmitoyltransferase 1B, acyl-CoA synthetase long chain family member 6, hydroxyacyl-CoA dehydrogenase and acyl-CoA dehydrogenase very long chain were gradually and significantly downregulated. The yellow module represented the specific physiological state of the normal heart at day 7, which is closely associated with the cGMP-PKG signaling pathway. The cGMP/PKG pathway is known to inhibit cardiac structural remodeling (28). All these results suggested heart remodeling following IR is a complex process, involving ECM, neural development, apoptosis and energy metabolism shift.
Hub genes were also identified in each module, including Src for the blue module, Dlg1 for the yellow module, Acly for the brown module, Ran for the green module, TP53 for turquoise module and Plk2 for red module. Src, upregulated in the heart 7 days following IR, is a non-receptor tyrosine kinase that serves roles in numerous biological process including cell adhesion, cell cycle and cell migration (29,30). In the heart, Src is activated by AngII and contributes to pathophysiology of cardiac remodeling and hypertrophy (23). It has been suggested that Src may be a candidate target for heart remodeling diseases (31). Dlg1, upregulated in the day 7 normal heart, encodes a multi-domain scaffolding protein, taking part in skeletogenesis, cytoskeleton organization and endothelia proliferation (32,33). Although there have not been any studies on its function in the heart, to the best of the authors' knowledge, it may be a target to improve heart function based on its function in other cell types. Acly, upregulated in the IR heart, is an ATP citrate lyase which is responsible for the synthesis of cytosolic acetyl-CoA (34). Acly-mediated dimethyl α-ketoglutarate prevents autophagy of heart and reduces heart contractile performance (35). Acly may be an adaptive response in the IR heart and may be used for the inhibition of autophagy in the heart. Ran, a member of RAS superfamily, was upregulated in the day 2 IR heart. Although there have not been any studies on its function in the heart, to the best of the authors' knowledge, previous studies have suggested that Ran controls microtubule organization during the apoptotic process (36,37). Ran can perform a similar function in cardiomyocytes (38), which should to be studied in the future. TP53, gradually upregulated in the IR heart, is key for heart disease progression. A number of studies have suggested that TP53 serves an important role in heart remodeling and heart failure, and is responsible for suppression of hypoxia inducible factor 1-induced angiogenesis, apoptosis of cardiomyocytes and fatty acid metabolism (39–41). Plk2, upregulated in the day 7 heart, is a member of the serine/threonine protein kinases. Plk2 mediates antioxidant signaling and is essential for preventing p53-dependent necrotic cell death in cancer cells and so may possess therapeutic implications for heart diseases (42,43).
Taken together, the results of the current study revealed the functional modules for heart remodeling following IR using a WGCNA method and identified key genes in each module by protein-protein interaction network. The results demonstrated that heart remodeling following IR is a complex process. A number of biological processes and signaling pathways are involved. The results of the present study may provide useful targets for heart disease treatment.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- WGCNA
weighted gene co-expression network analysis
- PPI
protein-protein interaction network
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DEGs
differentially expressed genes
- AMI
acute myocardial infarction
- ECM
extracellular matrix
Funding
No funding was received.
Availability of data and materials
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
Study design: NG, NZ, LY and ZL; data analysis: JW, FL, YW and XC; manuscript preparation: NG. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.GBD 2015 Disease and Injury Incidence and Prevalence Collaborators: Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the global burden of disease study 2015. Lancet. 2016;388:1545–1602. doi: 10.1016/S0140-6736(16)31678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ojha N, Roy S, Radtke J, Simonetti O, Gnyawali S, Zweier JL, Kuppusamy P, Sen CK. Characterization of the structural and functional changes in the myocardium following focal ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2008;294:H2435–H2443. doi: 10.1152/ajpheart.01190.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aversano T, Aversano LT, Passamani E, Knatterud GL, Terrin ML, Williams DO, Forman SA, Atlantic Cardiovascular Patient Outcomes Research Team (C-PORT) Thrombolytic therapy vs primary percutaneous coronary intervention for myocardial infarction in patients presenting to hospitals without on-site cardiac surgery: A randomized controlled trial. Jama. 2002;287:1943–1951. doi: 10.1001/jama.287.15.1943. [DOI] [PubMed] [Google Scholar]
- 4.Sen CK, Khanna S, Roy S. Perceived hyperoxia: Oxygen-induced remodeling of the reoxygenated heart. Cardiovasc Res. 2006;71:280–288. doi: 10.1016/j.cardiores.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Roy S, Khanna S, Bickerstaff AA, Subramanian SV, Atalay M, Bierl M, Pendyala S, Levy D, Sharma N, Venojarvi M, et al. Oxygen sensing by primary cardiac fibroblasts: A key role of p21(Waf1/Cip1/Sdi1) Circ Res. 2003;92:264–271. doi: 10.1161/01.RES.0000056770.30922.E6. [DOI] [PubMed] [Google Scholar]
- 6.Roy S, Khanna S, Wallace WA, Lappalainen J, Rink C, Cardounel AJ, Zweier JL, Sen CK. Characterization of perceived hyperoxia in isolated primary cardiac fibroblasts and in the reoxygenated heart. J Biol Chem. 2003;278:47129–47135. doi: 10.1074/jbc.M308703200. [DOI] [PubMed] [Google Scholar]
- 7.McNamara MT, Higgins CB. Magnetic resonance imaging of chronic myocardial infarcts in man. AJR Am J Roentgenol. 1986;146:315–320. doi: 10.2214/ajr.146.2.315. [DOI] [PubMed] [Google Scholar]
- 8.Orn S, Manhenke C, Anand IS, Squire I, Nagel E, Edvardsen T, Dickstein K. Effect of left ventricular scar size, location, and transmurality on left ventricular remodeling with healed myocardial infarction. Am J Cardiol. 2007;99:1109–1114. doi: 10.1016/j.amjcard.2006.11.059. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 10.Maschietto M, Tahira AC, Puga R, Lima L, Mariani D, Bda Paulsen S, Belmonte-de-Abreu P, Vieira H, Krepischi AC, Carraro DM, et al. Co-expression network of neural-differentiation genes shows specific pattern in schizophrenia. BMC Med Genomics. 2015;8:23. doi: 10.1186/s12920-015-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y, Xing Y. Weighted gene co-expression network analysis of pneumocytes under exposure to a carcinogenic dose of chloroprene. Life Sci. 2016;151:339–347. doi: 10.1016/j.lfs.2016.02.074. [DOI] [PubMed] [Google Scholar]
- 12.Tian F, Zhao J, Fan X, Kang Z. Weighted gene co-expression network analysis in identification of metastasis-related genes of lung squamous cell carcinoma based on the cancer genome atlas database. J Thorac Dis. 2017;9:42–53. doi: 10.21037/jtd.2017.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy S, Khanna S, Kuhn DE, Rink C, Williams WT, Zweier JL, Sen CK. Transcriptome analysis of the ischemia-reperfused remodeling myocardium: Temporal changes in inflammation and extracellular matrix. Physiol Genomics. 2006;25:364–374. doi: 10.1152/physiolgenomics.00013.2006. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haendeler J, Berk BC. Angiotensin II mediated signal transduction. Important role of tyrosine kinases. Regul Pept. 2000;95:1–7. doi: 10.1016/S0167-0115(00)00133-6. [DOI] [PubMed] [Google Scholar]
- 24.McMullen JR, Shioi T, Zhang L, Tarnavski O, Sherwood MC, Kang PM, Izumo S. Phosphoinositide 3-kinase(p110alpha) plays a critical role for the induction of physiological, but not pathological, cardiac hypertrophy. Proc Natl Acad Sci USA. 2003;100:12355–12360. doi: 10.1073/pnas.1934654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raaf L, Noll C, Mel Cherifi H, Samuel JL, Delcayre C, Delabar JM, Benazzoug Y, Janel N. Myocardial fibrosis and TGFB expression in hyperhomocysteinemic rats. Mol Cell Biochem. 2011;347:63–70. doi: 10.1007/s11010-010-0612-5. [DOI] [PubMed] [Google Scholar]
- 26.Shanaki M, Hossein-Nezhad A, Meshkani R, Beigy M, Shirzad M, Pasalar P, Golmohammadi T. Effects of resveratrol on crosstalk between canonical B-Catenin/Wnt and FOXO pathways in coronary artery disease patients with metabolic syndrome: A case control study. Iran J Pharm Res. 2016;15:547–559. [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller-Hennessen M, Sigl J, Fuhrmann JC, Witt H, Reszka R, Schmitz O, Kastler J, Fischer JJ, Müller OJ, Giannitsis E, et al. Metabolic profiles in heart failure due to non-ischemic cardiomyopathy at rest and under exercise. ESC Heart Fail. 2017;4:178–189. doi: 10.1002/ehf2.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Wang D, Wang F, Shi S, Chen Y, Yang B, Tang Y, Huang C. Exendin-4 inhibits structural remodeling and improves Ca2+ homeostasis in rats with heart failure via the GLP-1 receptor through the eNOS/cGMP/PKG pathway. Peptides. 2017;90:69–77. doi: 10.1016/j.peptides.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Ziehr J, Sheibani N, Sorenson CM. Alterations in cell-adhesive and migratory properties of proximal tubule and collecting duct cells from bcl-2 -/- mice. Am J Physiol Renal Physiol. 2004;287:F1154–F1163. doi: 10.1152/ajprenal.00129.2004. [DOI] [PubMed] [Google Scholar]
- 30.Chetty S, Engquist EN, Mehanna E, Lui KO, Tsankov AM, Melton DA. A Src inhibitor regulates the cell cycle of human pluripotent stem cells and improves directed differentiation. J Cell Biol. 2015;210:1257–1268. doi: 10.1083/jcb.201502035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey P, Hawkes W, Hu J, Megone WV, Gautrot J, Anilkumar N, Zhang M, Hirvonen L, Cox S, Ehler E, et al. Cardiomyocytes sense matrix rigidity through a combination of muscle and non-muscle myosin contractions. Dev Cell. 2018;44(326–336):e323. doi: 10.1016/j.devcel.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laprise P, Viel A, Rivard N. Human homolog of disc-large is required for adherens junction assembly and differentiation of human intestinal epithelial cells. J Biol Chem. 2004;279:10157–10166. doi: 10.1074/jbc.M309843200. [DOI] [PubMed] [Google Scholar]
- 33.Rivera C, Simonson SJ, Yamben IF, Shatadal S, Nguyen MM, Beurg M, Lambert PF, Griep AE. Requirement for Dlgh-1 in planar cell polarity and skeletogenesis during vertebrate development. PLoS One. 2013;8:e54410. doi: 10.1371/journal.pone.0054410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elshourbagy NA, Near JC, Kmetz PJ, Wells TN, Groot PH, Saxty BA, Hughes SA, Franklin M, Gloger IS. Cloning and expression of a human ATP-citrate lyase cDNA. Eur J Biochem. 1992;204:491–499. doi: 10.1111/j.1432-1033.1992.tb16659.x. [DOI] [PubMed] [Google Scholar]
- 35.Mariño G, Pietrocola F, Kong Y, Eisenberg T, Hill JA, Madeo F, Kroemer G. Dimethyl α-ketoglutarate inhibits maladaptive autophagy in pressure overload-induced cardiomyopathy. Autophagy. 2014;10:930–932. doi: 10.4161/auto.28235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dallol A, Hesson LB, Matallanas D, Cooper WN, O'Neill E, Maher ER, Kolch W, Latif F. RAN GTPase is a RASSF1A effector involved in controlling microtubule organization. Curr Biol. 2009;19:1227–1232. doi: 10.1016/j.cub.2009.05.064. [DOI] [PubMed] [Google Scholar]
- 37.Moss DK, Wilde A, Lane JD. Dynamic release of nuclear RanGTP triggers TPX2-dependent microtubule assembly during the apoptotic execution phase. J Cell Sci. 2009;122:644–655. doi: 10.1242/jcs.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang X, Zhang D, Zhang H, Huang Y, Teng M. Corrigendum: Role of Ran-regulated nuclear-cytoplasmic trafficking of pVHL in the regulation of microtubular stability-mediated HIF-1α in hypoxic cardiomyocytes. Sci Rep. 2015;5:11307. doi: 10.1038/srep11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita T, Ishikawa Y. Apoptosis in heart failure. -The role of the β-adrenergic receptor-mediated signaling pathway and p53-mediated signaling pathway in the apoptosis of cardiomyocytes. Circ J. 2011;75:1811–1818. doi: 10.1253/circj.CJ-11-0025. [DOI] [PubMed] [Google Scholar]
- 40.Oka T, Morita H, Komuro I. Novel molecular mechanisms and regeneration therapy for heart failure. J Mol Cell Cardiol. 2016;92:46–51. doi: 10.1016/j.yjmcc.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 41.Teng H, Sui X, Zhou C, Shen C, Yang Y, Zhang P, Guo X, Huo R. Fatty acid degradation plays an essential role in proliferation of mouse female primordial germ cells via the p53-dependent cell cycle regulation. Cell Cycle. 2016;15:425–431. doi: 10.1080/15384101.2015.1127473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Ma W, Wang PY, Hurley PJ, Bunz F, Hwang PM. Polo-like kinase 2 activates an antioxidant pathway to promote the survival of cells with mitochondrial dysfunction. Free Radic Biol Med. 2014;73:270–277. doi: 10.1016/j.freeradbiomed.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto T, Wang PY, Ma W, Sung HJ, Matoba S, Hwang PM. Polo-like kinases mediate cell survival in mitochondrial dysfunction. Proc Natl Acad Sci USA. 2009;106:14542–14546. doi: 10.1073/pnas.0904229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.