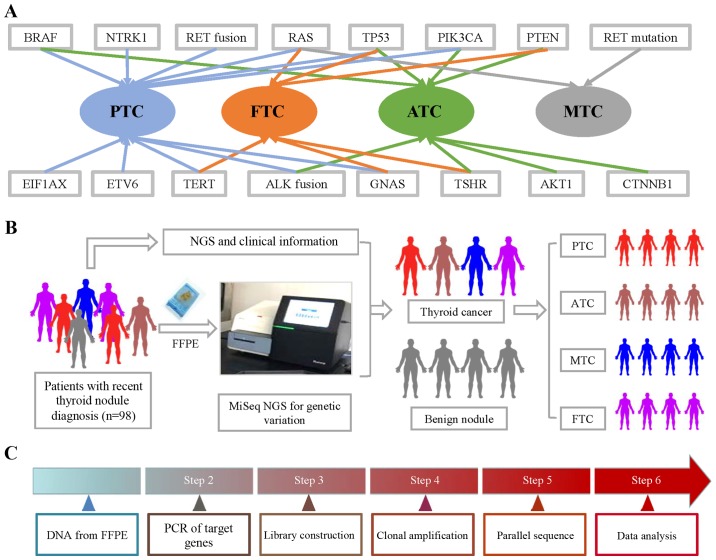
Figure 1.
Gene mutations and fusions in subtypes of TC and workflow of NGS. (A) BRAF, RAS, TERT, ETV6, EIF1AX, GNAS, PIK3CA, TP53 and NTRK1 mutations, as well as RET and ALK fusions, were found in PTC. BRAF, TERT, ALK fusion, GNAS, AKT1, PIK3CA, TP53 and PTEN were found in ATC. RAS, TERT, TSHR, GNAS, PENT and TP53 were found in FTC, while only RET and RAS mutations were found in MTC. (B) FFPE samples were obtained from 98 thyroid nodule patients, which was followed by CTC enumeration on NanoVelcro Chips. After collecting clinical information, we analyzed the correlation between pathological information and NGS results. (C) DNA from FFPE tissue was amplified for enrichment of target regions in a multiplex PCR reaction. Then, the library was prepared by ligating the PCR amplicons into platform-specific adapters and adding bar codes for specimen multiplexing. Finally, the library was enriched by clonal amplification (emPCR) and sequenced by massively parallel sequencing on the Ion Torrent PGM. The data analysis and variant calling were performed using bioinformatic pipelines followed by a custom SeqReporter algorithm for filtering and annotation of genetic variants. TC, thyroid cancer; NGS, next-generation sequencing; PTC, papillary thyroid cancer; ATC, anaplastic thyroid cancer; FTC, follicular thyroid cancer; MTC, medullary thyroid cancer; FFPE, formalin-fixed, paraffin-embedded.