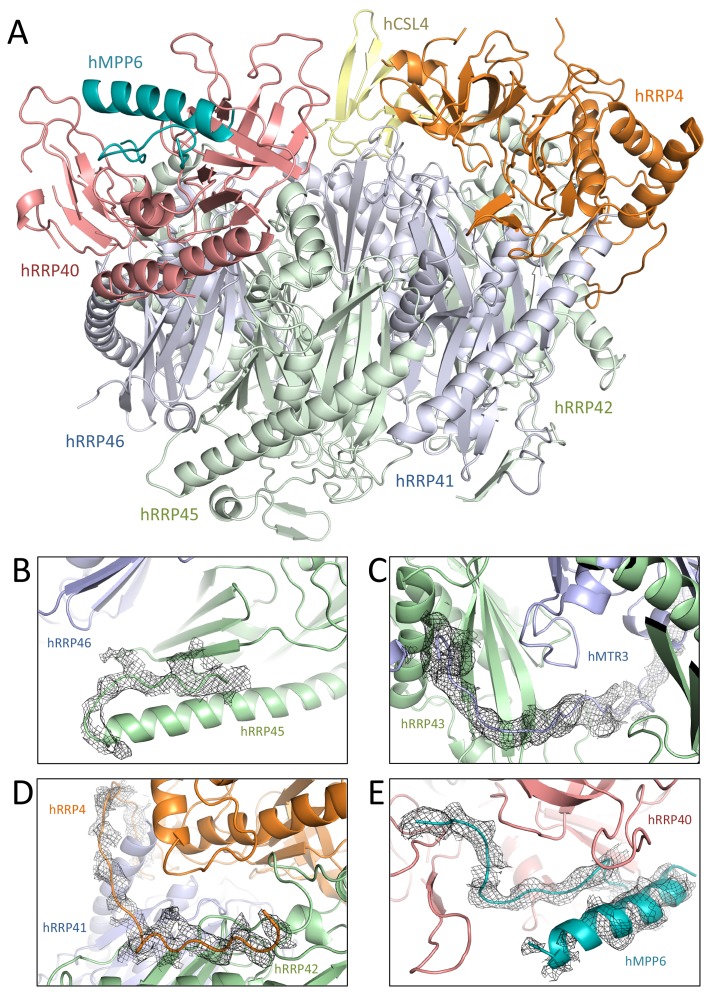
Figure 3. Human and yeast exosome cores: extensive similarities of the central scaffolds.
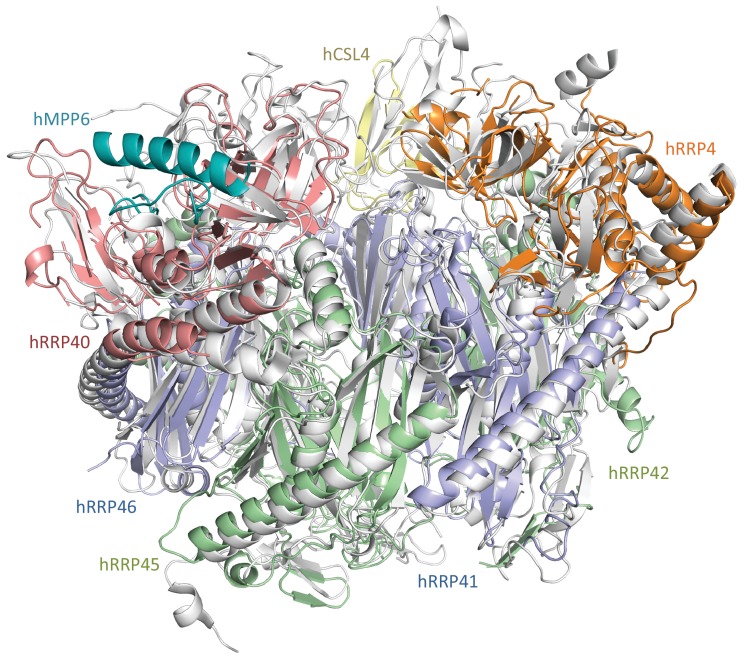
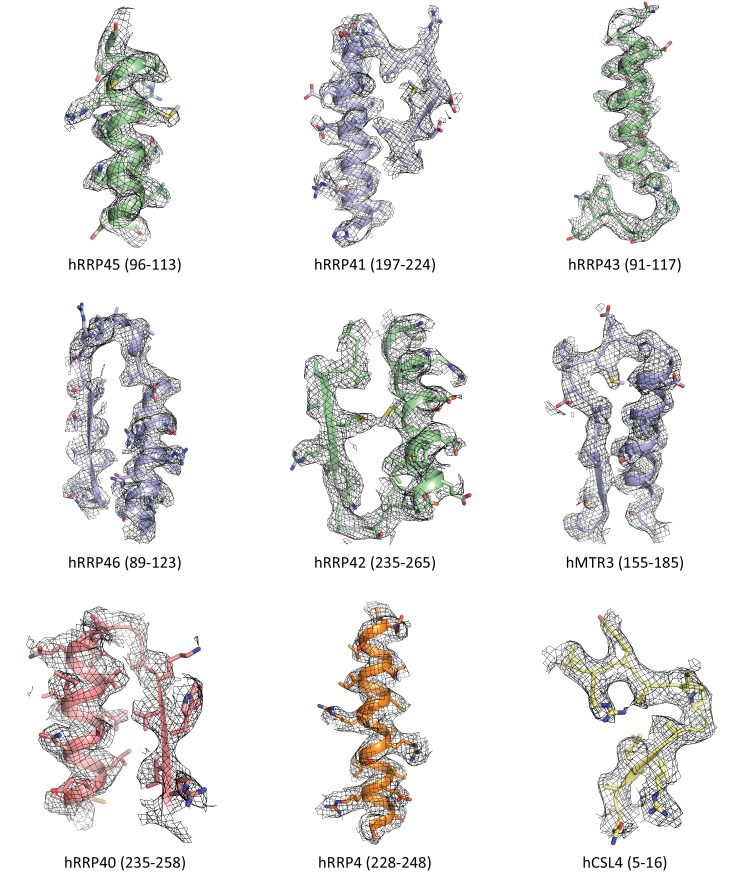
(A) Structure of the hEXO-9 barrel from the cryo-EM reconstruction bound to hMPP6 (teal). In hEXO-9, the cap proteins are shown in the same colors as in panel A and the base proteins are in light blue (for the Rrp41-like proteins hRRP41, hRRP46 and hMTR3) and light green (for the Rrp42-like proteins hRRP42, hRRP43 and hRRP45). The definition of Rrp41-like and Rrp42-like follows the original description in (Lorentzen et al., 2005). The superposition of the human and yeast scaffolds is shown in Figure 3—figure supplement 1. (B–D) Zoom-ins at specific regions of human hEXO-9 described in text, with cryo-EM density superposed. (E) Zoom-in at hMPP6 fragment bound to hRRP40, with cryo-EM density superposed.