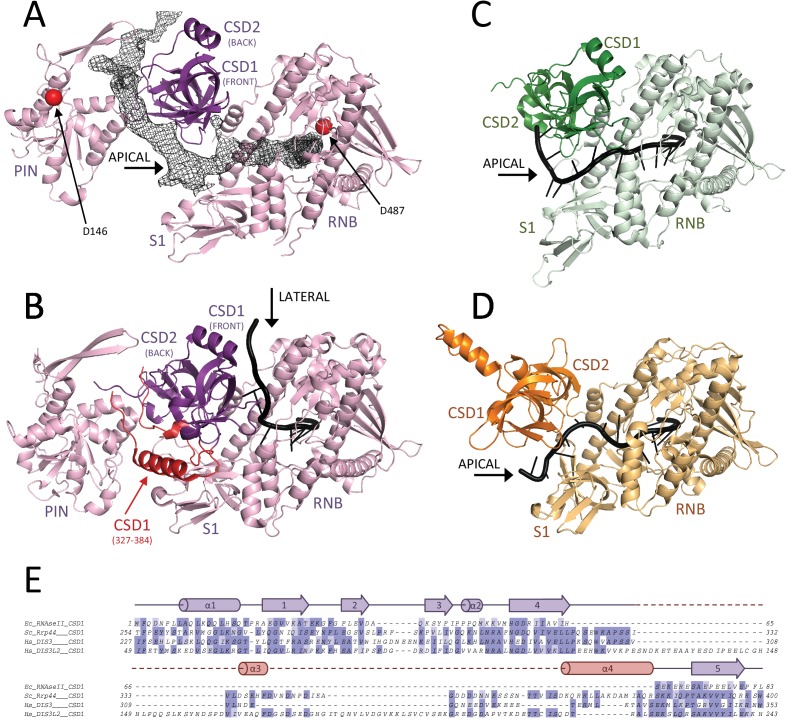
Figure 5. RNA binding to hDIS3 is more similar to hDIS3L2 and RNase II than to yRrp44.
(A) The structure of hDIS3 from the cryo-EM study is shown with the density corresponding to the bound RNA, segmented from the autorefined hEXO-10 map. (B) The structure of the S. cerevisiae orthologue yRrp44 in the open conformation with an RNA molecule accessing the RNB active site through the lateral entry. CSD1 region which impairs apical entry through the CSD1-S1 route is depicted in red (Makino et al., 2015). (C) The crystal structures of the paralogue mouse DIS3L2 (Faehnle et al., 2014) and (D) of the E. coli RNase II (Frazão et al., 2006). All structures are shown in the same orientation after optimal superposition of their RNB domains. In all panels the RNB and S1 domains form a rather rigid module and are shown in lighter colors, while the CSD1 and CSD2 domains are shown in darker colors. (E) Sequence alignment of the CSD1 domains from E. coli RNase II, S. cerevisiae yRrp44, human DIS3, and human DIS3L2. The secondary structure of the yRrp44 CSD1 region that impairs apical entry through the CSD1-S1 route is depicted in red.