 The replacement of the hydroxamic acid zinc-binding group in benzohydroxamic acid HDAC6 inhibitors by a trifluoromethyl ketone function leads to severe reduction in enzymatic and cellular activity.
The replacement of the hydroxamic acid zinc-binding group in benzohydroxamic acid HDAC6 inhibitors by a trifluoromethyl ketone function leads to severe reduction in enzymatic and cellular activity.
Abstract
Recent studies point towards the possible disadvantages of using hydroxamic acid-based zinc-binding groups in HDAC inhibitors due to e.g. mutagenicity issues. In this work, we elaborated on our previously developed Tubathian series, a class of highly selective thiaheterocyclic HDAC6 inhibitors, by replacing the benzohydroxamic acid function by an alternative zinc chelator, i.e., an aromatic trifluoromethyl ketone. Unfortunately, these compounds showed a reduced potency to inhibit HDAC6 as compared to their hydroxamic acid counterparts. In agreement, the most active trifluoromethyl ketone was unable to influence the growth of SK-OV-3 ovarian cancer cells nor to alter the acetylation status of tubulin and histone H3. These data suggest that replacement of the zinc-binding hydroxamic acid function with a trifluoromethyl ketone zinc-binding moiety within reported benzohydroxamic HDAC6 inhibitors should not be considered as a standard strategy in HDAC inhibitor development.
Introduction
The acetylation status of proteins is regulated by the subtle interplay between histone acetyltransferases (HATs) and histone deacetylases (HDACs). These enzymes respectively add or remove an acetyl group on lysine residues in specific proteins, thus influencing many important cell processes such as proliferation and protein degradation.1,2 In light of this extensive impact on cellular functions, it is clear that aberrant HDAC activity is an important actor in a vast amount of diseases, including different types of cancer, inflammation and neurodegenerative disorders.3–5 The HDAC family consists of 18 members (isozymes), which are divided into four classes: the zinc-dependent class I (HDAC1–3 and 8), class II (HDAC4–7 and 9–10) and class IV (HDAC11), and the NAD+-dependent class III (SIRT1–7).6,7 The inhibition of these enzymes by small molecules has proven to be an effective strategy in cancer therapy.3
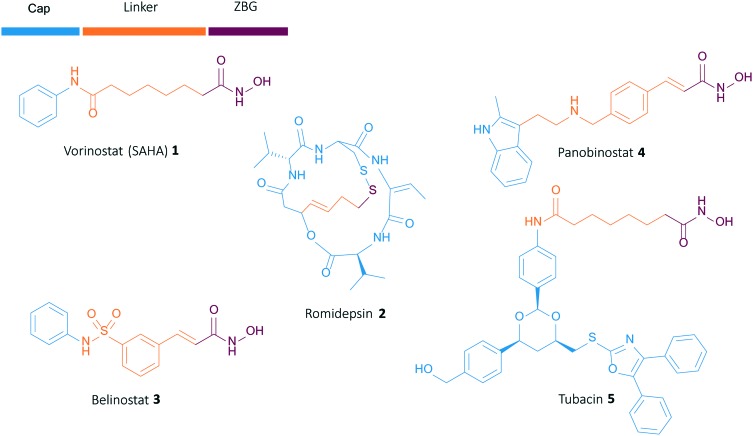
Most HDAC inhibitors consist of three parts that contribute to the overall biological effect: a ‘cap’-group, interacting with the enzyme surface, a linker unit, and a zinc-binding group (ZBG), which binds the zinc atom in the catalytic pocket. Currently, four HDAC inhibitors (HDACi's), i.e., Vorinostat 1, Romidepsin 2, Belinostat 3 and Panobinostat 4, are approved by the FDA as anticancer agents and are used in the clinic (Fig. 1). However, these inhibitors display a variety of side effects, possibly resulting from their non-selective character, meaning that they interfere with multiple zinc-dependent HDAC isozymes.8,9 To circumvent these problems, recent studies focus on the development of isoform-selective HDACi's. In particular, the selective inhibition of HDAC6 has proven to be of interest due to the cellular location and substrate specificity of this isozyme.10 The first selective HDAC6 inhibitor, Tubacin 5, showed good activity and selectivity, although its high lipophilicity and poor drug likeness hampered further applications.11 Since then, a variety of selective HDAC6 inhibitors has been designed, with the discovery of Tubastatin A 6 as an important milestone due to its excellent activity, selectivity and pharmacological profile.12–14 Our group recently developed a series of sulfur-containing Tubastatin A analogues, denoted as Tubathians 7, demonstrating an excellent activity and selectivity against HDAC6 in enzymatic and cellular assays, as well as desirable ADME-Tox properties.15,16 The most promising candidates within our series comprise a sulfoxide/sulfone-containing ‘cap’-group and an aromatic linker unit bearing a para-positioned hydroxamic acid functionality. This hydroxamic acid moiety acts as a powerful zinc chelator, making it the most widely used and the most effective zinc-binding group in HDACi development to date.17 However, recent literature points to the possible downsides of this particular ZBG, such as low bio-availability, toxicity due to non-specific metal binding, and mutagenicity.18–23 In light of these drawbacks, various other groups have been proposed as alternatives for the hydroxamic acid functionality, such as thiols, sulfonamides, boronic acids and many more, with rather mixed results.24
Fig. 1. Structure of reported HDAC inhibitors.
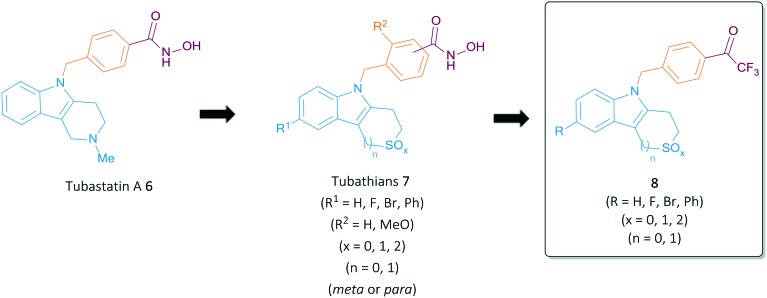
Recent work on HDAC6 inhibitors by our team focused on the optimization of the ‘cap’-group and linker unit of Tubathian analogues 7, keeping the ZBG constant.16 On the other hand, the present study aimed at replacing the potentially hazardous hydroxamic acid by another ZBG, namely a trifluoromethyl ketone (TFMK), in an attempt to further optimize this promising class of Tubathian HDAC6i's. The TMFK functionality has proven to be an effective metal chelator in various HDAC inhibitors, successfully replacing the hydroxamic acid group in for example Vorinostat 1.25 Inspired by these results, multiple trifluoromethyl ketone derivatives have been constructed in recent years, mainly containing an alkyl linker unit connected to a variety of ‘cap’-groups, such as tetrapeptides, 1,2,4-oxodiazoles and bisthiazoles.26–28 Furthermore, the use of an aromatic thiophene or furan linker resulted in a set of highly active and selective class II HDAC inhibitors.29,30 These literature results provided a clear rationale for the replacement of the hydroxamic acid within our previously developed HDAC6 inhibitors by a TFMK in order to further explore the structure–activity relationships (SAR) of this class of compounds (Fig. 2).
Fig. 2. Extended SAR of Tubathian analogues and target structures 8.
Results and discussion
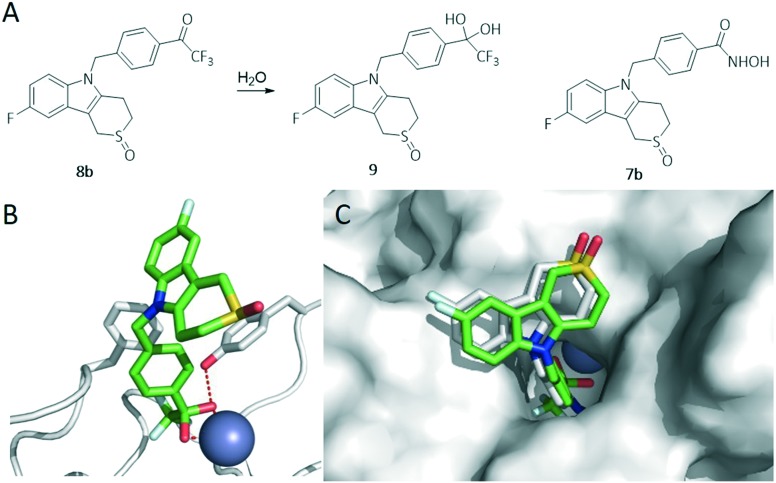
Prior to exploring the laboratory synthesis of novel Tubathian analogues 8 containing a trifluoromethyl ketone ZBG, in silico docking studies were performed using one of the envisioned compounds (8b) with respect to HDAC6 (Fig. 3). The trifluoromethyl ketone was docked as a hydrate 9, since this functional group exhibits its metal-chelating properties as such under physiological circumstances.24 The binding energy obtained for this molecule was comparable to that of the corresponding hydroxamic acid-containing Tubathian 7b (7.7 and 8.1 kcal mol–1, resp.). The ZBG of both compounds (7b and 9) interacted with the zinc atom, but the analogue with the trifluoromethyl ketone group also seemed to form a hydrogen bond with residue Tyr782. The positions of the linker group in the tubular access channel and the ‘cap’-group near the protein surface were comparable for both molecules, with no notable different interactions. This result suggests a possible inhibitory activity of the proposed compounds, justifying lab synthesis and further biological evaluation of these potential new HDAC6i's (for more information regarding these docking studies, see ESI†).
Fig. 3. A) Structures of docked compounds. B) Docking of compound 9 in catalytic domain CD2 of HDAC6 (hydrogen bonds are depicted in red). C) Overlap between hydrate 9 and the corresponding hydroxamic acid 7b.
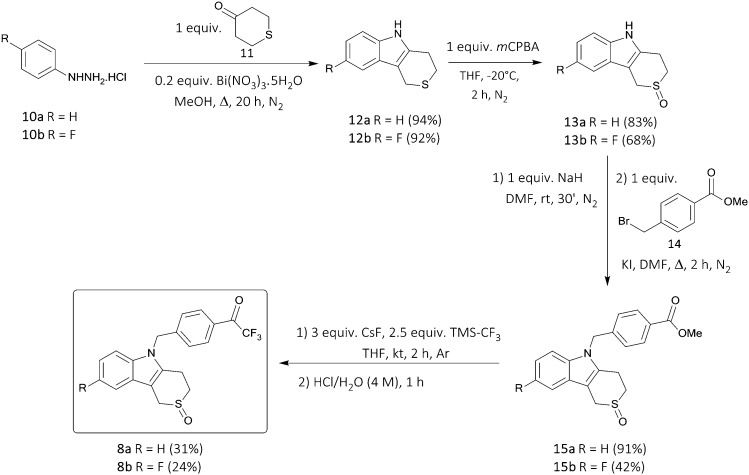
The synthesis of the proposed structures is based on our previous work regarding the preparation of Tubathians,15 starting with the formation of the tricyclic ‘cap’-group via a bismuth nitrate-catalyzed Fischer-indole synthesis using phenylhydrazine hydrochlorides 10 and tetrahydrothiopyranone 11 (Scheme 1). The sulfur atom present in the tricyclic structure was then selectively oxidized to a sulfoxide moiety using one equivalent of meta-chloroperbenzoic acid at low temperatures. Introduction of the aromatic linker unit was performed by N-deprotonation and subsequent nucleophilic substitution on methyl 4-(bromomethyl)benzoate 14.15,16 In the next step, the desired trifluoromethyl ketone ZBG was obtained by transformation of the methyl ester using Ruppert's reagent (TMSCF3), yielding the potential HDAC6 inhibitors 8 in moderate yields and high purity.31 In earlier experiments, the reaction of 4-bromo-(or 4-iodo-)benzyl bromide with the ‘cap’-group 13 was performed as well, after which the introduction of the trifluoromethyl ketone ZBG was attempted via either Grignard reaction or halogen–lithium exchange (using BuLi) with ethyl trifluoroacetate, albeit without any success. The final ester-to-CF3 ketone functional group interconversion step was also evaluated using sulfide- and sulfone-containing ‘cap’-groups (the S- and SO2-counterparts of structures 15), only leading to complex reaction mixtures. Nonetheless, since the sulfoxide analogues of the previously synthesized hydroxamic acid-containing Tubathians showed the best pharmacokinetic properties, no further attempts were made to obtain the sulfide and sulfone derivatives, and the biological potential of the unprecedented aromatic trifluoromethyl ketone structures 8 was assessed next.
Scheme 1. Synthesis of trifluoromethyl ketones 8.
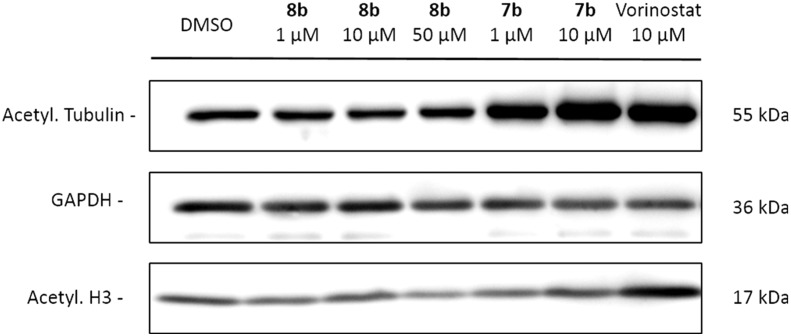
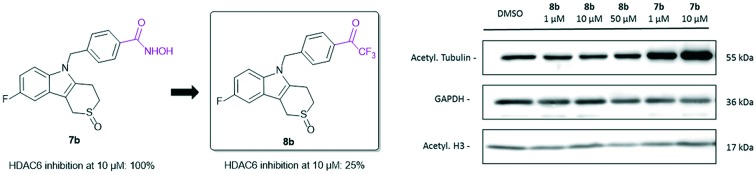
The two newly synthesized trifluoromethyl ketones 8a,b were first tested for their ability to inhibit HDAC6 on an enzymatic level. The percentage of HDAC6 inhibition using 10 μM of both compounds was observed to be 17 and 25%, respectively (see ESI†). This stands in high contrast to the effect of the hydroxamic acid analogue 7b, which gave a 100% inhibition of the same enzyme at this concentration, suggesting that the trifluoromethyl ketone ZBG exerts a lower activity than the potent hydroxamic acid. A drop in inhibitory activity has also been reported when comparing the hydroxamic acid with the trifluoromethyl ketone analogue of Vorinostat 1, confirming the superiority of the former functionality.25 The most active trifluoromethyl ketone 8b was then screened for its ability to inhibit HDAC enzymes on an cellular level by investigating the effect of the compound on the acetylation status of tubulin (an HDAC6 substrate) and histone H3 (nuclear HDACs substrate) in SK-OV-3 ovarian cancer cells via immunoblotting (Fig. 4, representative figure of three replicates is shown, see ESI†). As expected, no effect occurred when using 1 μM, but also no rise in acetylated tubulin was observed when using 10 and 50 μM, pointing to no (or very low) inhibition of the HDAC6 enzyme at these concentrations. This result contrasts with the effect observed when using the hydroxamic acid derivative 7b, which showed an increase in acetylated tubulin at a concentration as low as 1 μM. Both trifluoromethyl ketone 8b and hydroxamic acid 7b showed no increase in acetylated H3 at the concentrations used, indicating that no inhibition of nuclear HDACs occurred. The non-selective inhibitor Vorinostat 1 was deployed as a positive control and did result in an increase of the level of acetylated tubulin and H3 at a concentration of 10 μM. These immunoblotting results indicate that, although possessing some enzymatic HDAC6 activity, compound 8b is not capable of inhibiting the HDAC enzymes (both nuclear and cytoplasmatic) in cellular assays.
Fig. 4. Levels of acetylated tubulin and histone H3 in SK-OV-3 cells treated with different concentrations of 8b, 7b and control compound Vorinostat 1. GAPDH was used as a loading control.
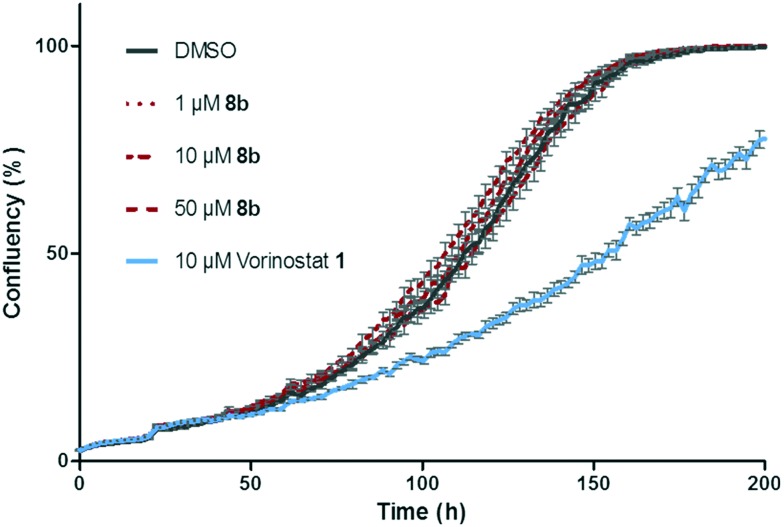
To confirm this, a functional cellular growth assay was performed using the trifluoromethyl ketone 8b (with Vorinostat 1 as a positive control). The inhibition of class I HDACs using Vorinostat 1 has been shown to suppress the growth of SK-OV-3 cancer cells, and the inhibition or knockdown of class II enzymes has been reported to have similar effects in various cancer cell lines.32–35 IncuCyte® ZOOM technology was used to measure the confluency of the SK-OV-3 cells in a 96-well plate, under control (0.1% DMSO) or treatment (as indicated) conditions (Fig. 5, representative graph of three replicates is shown, see ESI†). As expected, no impact was observed when using trifluoromethyl ketone 8b in concentrations ranging between 1 and 50 μM, confirming the immunoblotting data.
Fig. 5. Effects of trifluoromethyl ketone 8b on the proliferation of SK-OV-3 cells in different concentrations. Vorinostat 1 (10 μM) was used as a positive control. Mean + SEM is shown, N = 6.
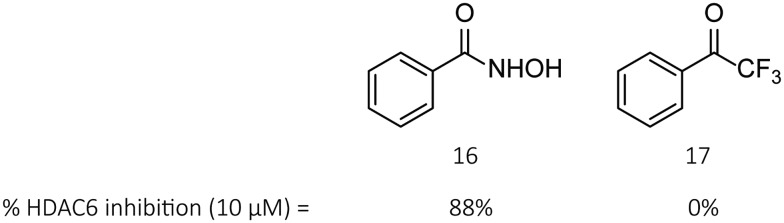
To further investigate whether or not the lack of HDAC6 inhibitory activity is directly linked to the change in the ZBG, we decided to compare the enzymatic HDAC6 inhibition of commercially available benzohydroxamic acid 16 and its trifluoromethyl ketone derivative 17 (Fig. 6) as a starting point, inspired by the literature information that potent and selective HDAC6 inhibition does not require a surface-binding motif.36 The percentages of HDAC6 inhibition at 10 μM by 16 and 17 were observed to be 88% and 0%, respectively, confirming that the replacement of the hydroxamic acid function by a trifluoromethyl ketone leads to a severe reduction of HDAC6 inhibitory efficacy. Benzohydroxamic acid 16, being a potent and selective ‘cap’-less HDAC6 inhibitor itself,36 comprises a vital part (linker unit and ZBG) of many distinguished HDAC6 inhibitors such as Tubastatin A, Nexturastat A and our Tubathian series. Consequently, due to the lack of activity in TFMKs 8 and 17, it can be concluded that a similar replacement in other benzohydroxamic acid-containing HDAC6 inhibitors will be pointless.
Fig. 6. Comparison of HDAC6 activity between benzohydroxamic acid 16 and its TFMK derivative 17.
The biological evaluation of the newly synthesized trifluoromethyl ketones proved to be inconsistent with the results of their in silico molecular docking. The docking studies showed no clear difference between the compounds containing a trifluoromethyl ketone 9 or hydroxamic acid 7b ZBG in both binding energy and pose. However, only the latter was shown to be a potent HDAC6 inhibitor in vitro. Molecular docking is widely regarded to be a helpful tool for determining the correct conformation of a ligand in a protein binding pocket, but the accuracy of its binding energy estimations is known to be rather limited.37 Furthermore, the scoring functions applied by docking methods have been reported to be particularly unreliable when the binding affinities for multiple highly similar ligands are being compared.38 This disadvantage emphasizes the need for experimental validation of the structure–activity relationships of HDAC inhibitors.
Recent HDAC-related research focuses on the development of isoform-selective HDAC inhibitors, mostly applying the hydroxamic acid function as the zinc-binding group.39 Although the use of aromatic linker units is known to induce class II HDAC selectivity, as indicated by the success of benzohydroxamic acid-based HDAC6 inhibitors, only two phenyl trifluoromethyl ketones have been designed so far.25,29 One of these compounds showed moderate inhibition of HDAC6 with an IC50-value of 2.5 μM in an enzymatic assay, but was not tested in a cellular context.29 The second compound has only been evaluated in a nuclear HDAC-extract mainly containing HDAC1 and 2, showing no activity.25 These data are in line with the lack of activity we observed in our newly developed trifluoromethyl ketone-containing Tubathians 8 and in the trifluoromethyl ketone derivative of benzohydroxamic acid 17. In contrast, the use of an aromatic thiophene linker unit has been reported to result in class II selective HDACi's, exerting low nanomolar activity against HDAC6.29 Nonetheless, these compounds also failed to induce cellular effects due to enzymatic reduction of the TFMK function to the inactive alcohol by carbonyl reductases present in the cells.40 The susceptibility of (aromatic) trifluoromethyl ketones to enzymatic transformations in a cellular environment, can, next to the poor enzymatic activity, possibly contribute to the ineffectiveness of compound 8b in SK-OV-3 ovarian cancer cells as well.
Conclusion
In conclusion, the replacement of the hydroxamic acid ZBG in benzohydroxamic HDAC6 selective inhibitors with a trifluoromethyl ketone, although being an effective strategy for non-selective inhibitors such as Vorinostat 1, appears to be detrimental with regard to enzymatic and cellular activity. These data suggest that, for the development of HDAC6 selective inhibitors incorporating an alternative zinc-binding group, the promising results of a structurally distinct inhibitor with a specific zinc chelator do not guaranty success when transferred to a new linker and ‘cap’-group.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
The authors declare no competing interests.
Supplementary Material
Acknowledgments
The Authors are indebted to the Ghent University Special Research Fund (BOF) for financial support.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8md00107c
References
- Dokmanovic M., Clarke C., Marks P. A. Mol. Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- Kim H.-J., Bae S.-C. Am. J. Transl. Res. 2011;3:166–179. [PMC free article] [PubMed] [Google Scholar]
- Bolden J. E., Peart M. J., Johnstone R. W. Nat. Rev. Drug Discovery. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L., Canettieri G., Infante P., Greco A., Gulino A. Biochim. Biophys. Acta, Rev. Cancer. 2011;1815:241–252. doi: 10.1016/j.bbcan.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Shein N. A. A., Shohami E. Mol. Med. 2011;17:448–456. doi: 10.2119/molmed.2011.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruijter A. J. M., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. P. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti I. V., Lee Y.-M., Goodson H. V. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Karagiannis T. C., El-Osta A. Leukemia. 2007;21:61–65. doi: 10.1038/sj.leu.2404464. [DOI] [PubMed] [Google Scholar]
- Thomas E. A. Mol. Neurobiol. 2009;40:33–45. doi: 10.1007/s12035-009-8067-y. [DOI] [PubMed] [Google Scholar]
- Aldana-Masangkay G. I., Sakamoto K. M. J. Biomed. Biotechnol. 2011;2011:875824. doi: 10.1155/2011/875824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. C., Hong R., Schreiber S. L. J. Am. Chem. Soc. 2003;125:5586–5587. doi: 10.1021/ja0341440. [DOI] [PubMed] [Google Scholar]
- Butler K. V., Kalin J., Brochier C., Vistoli G., Langley B., Kozikowski A. P. J. Am. Chem. Soc. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalin J. H., Butler K. V., Akimova T., Hancock W. W., Kozikowski A. P. J. Med. Chem. 2012;55:639–651. doi: 10.1021/jm200773h. [DOI] [PubMed] [Google Scholar]
- Woan K. V., Lienlaf M., Perez-Villaroel P., Lee C., Cheng F., Knox T., Woods D. M., Barrios K., Powers J., Sahakian E., Wang H. W., Canales J., Marante D., Smalley K. S. M., Bergman J., Seto E., Kozikowski A., Pinilla-Ibarz J., Sarnaik A., Celis E., Weber J., Sotomayor E. M., Villagra A. Mol. Oncol. 2015;9:1447–1457. doi: 10.1016/j.molonc.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vreese R., Verhaeghe T., Desmet T., D'hooghe M. Chem. Commun. 2013;49:3775–3777. doi: 10.1039/c3cc41422a. [DOI] [PubMed] [Google Scholar]
- De Vreese R., Depetter Y., Verhaeghe T., Desmet T., Benoy V., Haeck W., Van Den Bosch L., D'hooghe M. Org. Biomol. Chem. 2016;14:2537–2549. doi: 10.1039/c5ob02625c. [DOI] [PubMed] [Google Scholar]
- Wagner F. F., Weiwer M., Lewis M. C., Holson E. B. Neurotherapeutics. 2013;10:589–604. doi: 10.1007/s13311-013-0226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strevel E. L., Siu L. L. Eur. J. Cancer. 2009;45:318–331. doi: 10.1016/S0959-8049(09)70046-2. [DOI] [PubMed] [Google Scholar]
- Munakata K., Mochida H., Kondo S., Suzuki Y. J. Pharmacobio-Dyn. 1980;3:557–561. doi: 10.1248/bpb1978.3.557. [DOI] [PubMed] [Google Scholar]
- Johnson D., Walmsley R. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2013;751:96–100. [Google Scholar]
- Shen S., Kozikowski A. P. ChemMedChem. 2016;11:15–21. doi: 10.1002/cmdc.201500486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Isobe M. Cancer Res. 1990;50:4300–4307. [PubMed] [Google Scholar]
- Groutas W. C., Giri P. K., Crowley J. P., Castrisos J. C., Brubaker M. J. Biochem. Biophys. Res. Commun. 1986;141:741–748. doi: 10.1016/s0006-291x(86)80235-2. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Miyata N. Curr. Med. Chem. 2005;12:2867–2880. doi: 10.2174/092986705774454706. [DOI] [PubMed] [Google Scholar]
- Frey R. R., Wada C. K., Garland R. B., Curtin M. L., Michaelides M. R., Li J., Pease L. J., Glaser K. B., Marcotte P. A., Bouska J. J., Murphy S. S., Davidsen S. K. Bioorg. Med. Chem. Lett. 2002;12:3443–3447. doi: 10.1016/s0960-894x(02)00754-0. [DOI] [PubMed] [Google Scholar]
- Lai J.-I., Leman L. J., Ku S., Vickers C. J., Olsen C. A., Montero A., Ghadiri M. R., Gottesfeld J. M. Bioorg. Med. Chem. Lett. 2017;27:3289–3293. doi: 10.1016/j.bmcl.2017.06.027. [DOI] [PubMed] [Google Scholar]
- Muraglia E., Altamura S., Branca D., Cecchetti O., Ferrigno F., Orsale M. V., Palumbi M. C., Rowley M., Scarpelli R., Steinkuhler C., Jones P. Bioorg. Med. Chem. Lett. 2008;18:6083–6087. doi: 10.1016/j.bmcl.2008.09.076. [DOI] [PubMed] [Google Scholar]
- Gong C.-J., Gao A.-H., Zhang Y.-M., Su M.-B., Chen F., Sheng L., Zhou Y.-B., Li J.-Y., Li J., Nan F.-J. Eur. J. Med. Chem. 2016;112:81–90. doi: 10.1016/j.ejmech.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Jones P., Bottomley M. J., Carfi A., Cecchetti O., Ferrigno F., Lo Surdo P., Ontoria J. M., Rowley M., Scarpelli R., Schultz-Fademrecht C., Steinkuhler C. Bioorg. Med. Chem. Lett. 2008;18:3456–3461. doi: 10.1016/j.bmcl.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Ontoria J. M., Altamura S., Di Marco A., Ferrigno F., Laufer R., Muraglia E., Palumbi M. C., Rowley M., Scarpelli R., Schultz-Fademrecht C., Scrafini S., Steinkuhler C., Jones P. J. Med. Chem. 2009;52:6782–6789. doi: 10.1021/jm900555u. [DOI] [PubMed] [Google Scholar]
- Singh R. P., Cao G., Kirchmeier R. L., Shreeve J. N. M. J. Org. Chem. 1999;64:2873–2876. doi: 10.1021/jo982494c. [DOI] [PubMed] [Google Scholar]
- Chen M.-Y., Liao W. S. L., Lu Z., Bornmann W. G., Hennessey V., Washington M. N., Rosner G. L., Yu Y., Ahmed A. A., Bast Jr R. C. Cancer. 2011;117:4424–4438. doi: 10.1002/cncr.26073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T., Bradner J. E., Wong J., Chauhan D., Richardson P., Schreiber S. L., Anderson K. C. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8567–8572. doi: 10.1073/pnas.0503221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-S., Lim K.-H., Guo X., Kawaguchi Y., Gao Y., Barrientos T., Ordentlich P., Wang X.-F., Counter C. M., Yao T.-P. Cancer Res. 2008;68:7561–7569. doi: 10.1158/0008-5472.CAN-08-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu W., Yan H., Sun Z., Tang Y. Zhongguo Zhongliu Linchuang. 2013;40:698–701. [Google Scholar]
- Wagner F. F., Olson D. E., Gale J. P., Kaya T., Weiwer M., Aidoud N., Thomas M., Davoine E. L., Lemercier B. C., Zhang Y.-L., Holson E. B. J. Med. Chem. 2013;56:1772–1776. doi: 10.1021/jm301355j. [DOI] [PubMed] [Google Scholar]
- Wang Z., Sun H., Yao X., Li D., Xu L., Li Y., Tian S., Hou T. Phys. Chem. Chem. Phys. 2016;18:12964–12975. doi: 10.1039/c6cp01555g. [DOI] [PubMed] [Google Scholar]
- Ramirez D., Caballero J. Int. J. Mol. Sci. 2016;17:1–15. doi: 10.3390/ijms17040525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vreese R., D'hooghe M. Eur. J. Med. Chem. 2017;135:174–195. doi: 10.1016/j.ejmech.2017.04.013. [DOI] [PubMed] [Google Scholar]
- Scarpelli R., Di Marco A., Ferrigno F., Laufer R., Marcucci I., Muraglia E., Ontoria J. M., Rowley M., Serafini S., Steinkuhler C., Jones P. Bioorg. Med. Chem. Lett. 2008;18:6078–6082. doi: 10.1016/j.bmcl.2008.10.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.