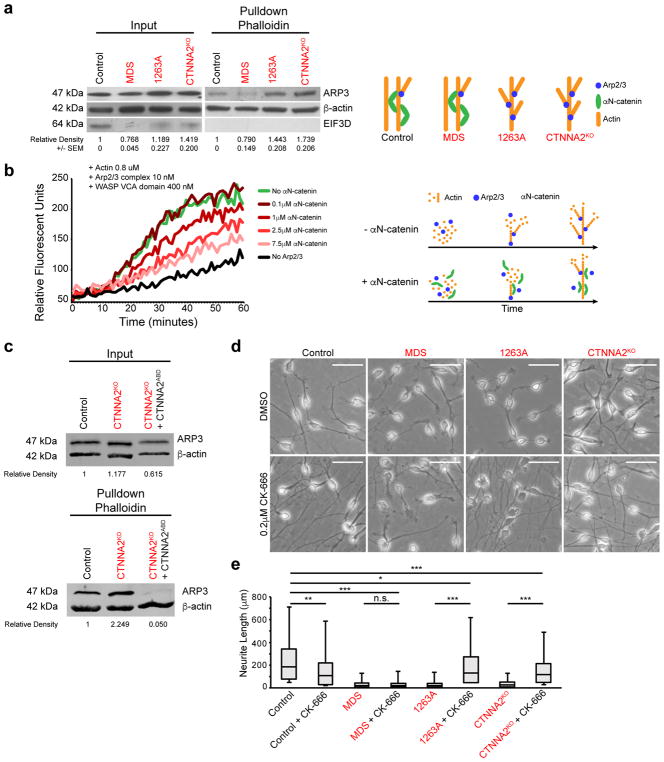
Figure 4. αN-catenin represses Arp2/3-actin association and polymerization.
(a) Elevated association between affinity-purified F-actin and ARP2/3 in CTNNA2-mutant cells. Input: basal expression. F-actin pulldown with phalloidin. Exaggerated ARP3 band in 1263A and knockout lines, compared MDS or Control line. EIF3D (does not bind actin) as a control for pulldown specificity. Cropped images shown. Repeated in triplicate, quantification and SEM below. Schematic depicts model (right).
(b) Actin polymerization assay showing dose-dependent inhibition of Arp2/3 + VCA domain of WASP mediated actin polymerization by αN-catenin, measured by relative fluorescent units. In the absence of Arp2/3 (black) there was minimal polymerization, but when Arp2/3 + VCA was added (green) polymerization increased substantially, which was reversed upon dose escalation of αN-catenin. Repeated in triplicate. Schematic depicts model (right).
(c) Loss of association between F-actin and ARP2/3 in CTNNA2KO cells expressing the actin binding domain (ABD) of αN-catenin. Input: basal expression of ARP3 and Actin. F-actin pulldown with phalloidin. Exaggerated ARP3 band in knockout neurons, and loss of ARP3 in ABD-expressing knockout neurons, compared with band in Control line. Quantification by relative fluorescence of ARP3 to Actin. Cropped images shown. Repeated in duplicate.
(d) ARP2/3 inhibition by CK-666 rescued neurite length defect in CTNNA2 mutant neurons, but not Control and MDS mutant neurons. Scale bar 50 μm.
(e) Quantification of (d) with top and bottom quartiles and Median displayed. Whiskers represent the minimum and maximum values observed in the dataset. Repeated in three independent iPSC clones per patient or three CTNNA2KO clones, 442 cells scored.*, ** and ***, significant P (see Statistics and Reproducibility).