Abstract
DNA double-strand breaks (DSBs) are the most cytotoxic DNA lesions, and up to 90% of DSBs require repair by non-homologous end joining (NHEJ). Functional and genomic analyses of patient-derived melanomas revealed that PTEN loss is associated with NHEJ deficiency. In PTEN-null melanomas PTEN complementation rescued the NHEJ defect; conversely suppression of PTEN compromised NHEJ. Mechanistic studies revealed that PTEN promotes NHEJ through direct induction of expression of XRCC4-like factor (NHEJ1/XLF) which functions in DNA end bridging and ligation. PTEN was found to occupy the NHEJ1 gene promoter and to recruit the histone acetyltransferases, PCAF and CBP, inducing XLF expression. This recruitment activity was found to be independent of its phosphatase activity but dependent on K128, a site of regulatory acetylation on PTEN. These findings define a novel function for PTEN in regulating NHEJ double-strand break repair, and therefore may assist in the design of individualized strategies for cancer therapy.
Introduction
DNA double-strand breaks (DSBs) are the most deleterious of DNA lesions and the majority (up to 90%) of DNA DSBs are repaired by end joining (1). The non-homologous end joining (NHEJ) pathway is a rapid process involving the Ku70-Ku80 heterodimer, DNA protein kinase (DNA-PK), and the XLF, XRCC4, DNA ligase IV complex. NHEJ is active in all phases of the cell cycle and is exclusively relied upon in G0 and G1, and so regulation of this pathway is paramount. Aberrant expression of the core NHEJ components confers dramatic cellular phenotypes: loss of Ku70 or Ku80 is lethal in human cells (2, 3), expression loss or pharmacological inhibition of DNA-PK produces severe defects in NHEJ (4) and loss of XLF expression yields marked NHEJ deficiency (5, 6).
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is a key tumor suppressor antagonizing oncogenic Protein Kinase B (AKT) and phosphoinositide 3-kinase (PI3K) signaling at the cell membrane and is frequently mutated or lost in human cancers. PTEN alteration has been reported in up to 15% of human melanomas (7), in 40% of melanoma cell lines (8), in 67% of uterine carcinomas (9), 49% of prostate carcinomas (10), and 38% of glioblastomas (11). PTEN knockout combined with expression of a BRAF V600E variant is sufficient for melanoma development in a mouse model (12). Additionally, nuclear PTEN was first identified after the observation that PTEN knockout in mouse embryonic fibroblasts yields chromosomal instability, and as a potential explanation it was proposed that PTEN regulates the HDR pathway via Rad51 (13). However, subsequent studies suggested that PTEN loss might not result in in altered RAD51 expression(14). Nonetheless, the possibility remained that PTEN may still have a role in DNA repair, as several recent studies reported that PTEN is a nuclear protein impacting sensitivity to ionizing radiation (IR) (14–16) and other genotoxic stresses (14, 17) as well as genome integrity (18–21).
Seeking to develop new targeted therapies for melanomas, we examined a panel of patient-derived human melanomas, with known variable levels of radiosensitivity (22), for NHEJ capacity using a host cell reactivation reporter assay, with the goal of establishing potential genetic correlations by reference to comprehensive whole exome sequencing (23, 24) and gene expression data (25) that are available for these melanomas. We report here the finding that patient derived melanomas deficient in PTEN show reduced NHEJ activity, and we elucidate a pathway by which PTEN regulates NHEJ through XLF. In patient-derived melanomas null for PTEN, PTEN complementation was consistently found to restore NHEJ activity, whereas PTEN suppression by siRNAs in PTEN wild-type melanoma cells yielded decreased NHEJ. By analysis of gene expression patterns, we identified a strong and specific association between PTEN levels and expression of the NHEJ factor, XLF, which plays an essential role in NHEJ via complex formation with XRCC4 and DNA ligase IV(5). Manipulation of PTEN expression in the melanomas established a direct link at the transcriptional level between PTEN and XLF expression. Functionally, we show that restoration of XLF expression in PTEN null melanomas reestablishes efficient NHEJ activity, and loss of XLF in wild type melanomas compromises NHEJ, supporting regulation of XLF by PTEN as a key regulatory point for the NHEJ pathway. Further, co-immunoprecipitation (co-IP) and chromatin immunoprecipitation (ChIP) experiments reveal physical interaction between PTEN and the transcriptional co-activators and histone acetyltransferases, PCAF and CBP, and place PTEN, PCAF and CBP at the XLF promoter in a PTEN-dependent manner. Notably, this effect of PTEN was found to be dependent on residue K128, a site of regulatory acetylation, but independent of PTEN’s phosphatase activity. Altogether, we present mechanistic evidence that PTEN regulates NHEJ activity through XLF via a novel pathway of PTEN-mediated epigenetic activation of the XLF promoter and identify PTEN loss as a potential biomarker for impaired NHEJ.
Materials and Methods
Cell culture
xrs6 Chinese hamster ovary cells deficient in Ku80 and the xrs6+Ku80 complemented cells were previously described (26). Both xrs6 and xrs6+Ku80 cells were cultured in Ham’s F12 medium + 10% FBS + 1X Pen/Strep. DLD1 and DLD1 BRCA2 −/− (Horizon Discovery) were cultured in DMEM medium + 10%FBS + 1X Pen/Strep. U2OS EJ5 were cultured in DMEM medium + 10% FBS + 1X Pen/Strep. Primary Melanoma cultures were maintained in OPTIMEM media with 5% FBS and 1% Pen/Strep. U251 cells have been previously described(27) and were cultured in DMEM+ 10% FBS 0.5 mg/ml G418 and 10 μg/ml blasticidin and PTEN expression was induced by doxycycline at 1 μg/ml. Parental HCT116 and PTEN −/− HCT116 cells were obtained from horizon discovery. Human melanocytes (Lonza) were cultured using the MGM-4 Melanocyte Growth Bullet Kit (Lonza). All cell lines tested negative for mycoplasma. Cell lines were obtained from the original publishing labs, and primary melanoma cells were obtained from the Yale Melanoma Specimen Research Core.
PTEN retroviral production and infection
pBABE-PURO PTEN, pBABE-PURO PTEN C124S, pBABE-PURO PTEN G129E and pBABE-PURO Empty Vector were obtained from Addgene and confirmed by Sanger sequencing. pBABE PURO PTEN K128R was created by site directed mutagenesis of PBABE-PURO PTEN using the Q5 Site-Directed Mutagenesis Kit (NE BioLabs) per manufacturer’s protocol using the primers: 5′AAAGCTGGAAGGGGACGAACT 3′ and 5′ACAGTGAATTGCTGCAAC 3′. To produce retroviral particles, 2.5 × 106 293FT cells were plated in a 15 cm dish 24 h before transfection and then transfected with 20 μg transfer vector, 18 μg pUMVC and 2 μg pCMV-VSVG using Lipofectamine 2000. Supernatant was collected 48 and 72 h later, filtered using a 0.45 μM filter, and combined. YUGEN8 and YUROL PTEN null melanoma cells were seeded to 20% confluence in a 10 cm dish 24 h before infection, and subsequently infected with 3.5 mL of filtered viral supernatant and 8 μg/mL polybrene for 4 h. The infection was repeated 24 h later. An additional 24 h after the second infection, cells were selected with puromycin at a concentration of 1 μg/mL. PTEN expression was confirmed by western blot.
Antibodies used for western blots
PTEN (sc7974, Santa Cruz Biotechnology), RAD51 (sc-8349, Santa Cruz Biotechnology), XLF (#2854, Cell Signaling Technologies), Vinculin (ab12058, Abcam), DNA-PK (#4602, Cell Signaling Technologies), KU80 (556429, BD Transduction Laboratories), XRCC4 (611506, BD Transduction Laboratories), PCAF (C14G9, Cell Signaling Technologies), CBP(D6C5, Cell Signaling Technologies), BRCA2 (Ab1, Millipore) phospho-AKT Ser473 (D9E, Cell Signaling Technologies), pan-AKT(11E7, Cell Signaling Technologies), B-Actin HRP ( 60008, Protein Tech).
Luciferase DNA Repair Reporter Assays
Luciferase assays for NHEJ have been previously described(28). Briefly, to assay NHEJ capacity, pGL3 control (Promega) was linearized with HindIII (NE Biolabs) and the double-strand break was confirmed by gel electrophoresis. 24 h before transfection, the melanoma cell populations were seeded at a density of 7×104 cells per well in 12-well format. 1 μg of linearized plasmid was transfected into the cells along with 50 ng pCMV-RL (Promega) as a transfection control using Lipofectamine 2000 (Life Technologies). In parallel, cells were transfected with 1 μg uncut pGL3-Control and 50 ng pCMV-RL as a positive control. Luciferase activity was assayed using the Dual Luciferase Reporter Assay Kit (Promega) 24 h after transfection. NHEJ activity was calculated by normalizing each transfection to the Renilla luciferase transfection efficiency control, and then normalizing cut pGL3-Control to uncut pGL3-Control to calculate percent reactivation by NHEJ. Data are presented as relative repair efficiencies, where percent reactivation from the experimental condition is normalized to the control condition.
Comet Assays
Cells were collected at the stated time points after 5 Gy IR treatment and interrogated for the presence of DNA DSBs by neutral comet assay (Trevigen) per the manufacturer’s protocol, as previously described (29) For comet assays in the context of DNA-PK inhibition, cells were treated with 10 μM DNA-PK inhibitor NU7441 (Selleck Chemicals) or DMSO for 24 h and then irradiated and collected 24 h after IR for the comet assay.
Immunofluorescence Imaging
Cells were fixed at indicated times after IR treatment and were stained with rabbit anti-γH2AX antibody (#9718, Cell Signaling Technology) and with 100 mg/ml DAPI (Sigma). Images were captured using an Axiovert 200 microscope (Carl Zeiss Micro Imaging, Inc.). Images were analyzed by counting foci per nucleus using Cell Profiler software.
siRNA Knockdown
Primary melanoma cultures were transfected in 10 cm dishes to a final concentration of 20 nM for siPTEN (ON-TARGETplus PTEN siRNA, GE Dharmacon), siXLF (ON-TARGETplus NHEJ1 siRNA, GE Dharmacon), siXRCC4 (ON-TARGETplus XRCC4 siRNA, GE Dharmacon), siXRCC5 (ON-TARGETplus XRCC5 siRNA, GE Dharmacon), siBRCA2 (ON-TARGETplus BRCA2 siRNA, GE Dharmacon), siRAD51 (ON-TARGETplus RAD51 siRNA, GE Dharmacon) and scrambled siRNA control (Negative Control siRNA duplex, Qiagen 1027310) with Dharmafect 2 (GE Dharmacon) per manufacturer’s protocol.
Clonogenic survival assays
Patient-derived melanoma cultures or melanoma cell lines growing at 60–70% confluence were treated with IR in 10 cm dishes in triplicate and then plated at 100 to 4800 cells/well in 6-well plates. Cells were cultured for 1 to 2 weeks until well-defined colonies were formed. For siRNA experiments, cells were treated with IR at 72 h after siRNA transfection. For quantification of colonies, cells were briefly permeabilized with 0.9% saline solution and then stained with a crystal violet solution in 80% methanol.
U2OS EJ5 reporter assays
Briefly, 1×106 cells were transfected in triplicate with 4 μg pI-SceI using the Amaxa Nucleofector II and Nucleofection Kit V (Lonza) per manufacturer’s protocol. 72 h after transfection cells were analyzed for GFP expression using flow cytometry, and the data were analyzed using FlowJo software.
Microarray Data Analysis
The Skin Cancer Microarray data from the Yale Melanoma Gene Expression Cohort has been previously described (Parisi et al., 2012). The data set is available through the Melagrid resource (http://melagrid.org).
Analysis of Publicly Available TCGA Sequencing Data
Processed publicly available data were downloaded via cBioPortal.
Statistical Analyses
Statistical analyses were conducted using Prism 6.0 (Graph Pad).
Reverse Transcriptase Quantitative PCR analysis (qRT-PCR)
Total RNA was prepared using the RNAeasy Miniprep Kit (Qiagen). 1 μg of RNA was used in the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The resulting cDNA was diluted 1:5 and combined with primers and Power SYBR Green PCR Master Mix (Applied Biosystems/Life Technologies). Plates were spun down prior to analysis. The Mx3000p real time PCR system (Stratagene) was used to monitor fluorescence intensity in real-time to allow quantitative comparisons. Ct values were normalized to GAPDH and relative expression was calculated using the −ΔΔCt method.
qPCR primer sequences
NHEJ1/XLF mRNA F: 5′ GGCCAAGGTTTTTATCACCAAGC 3′
R: 5′ TGGGCGAAGGAGATTATCCAAAT 3′
PTEN mRNA F: 5′ TGGATTCGACTTAGACTTGACCT 3′
R: 5′ GGTGGGTTATGGTCTTCAAAAGG 3′
GAPDH mRNA F: 5′ GGAGCGAGATCCCTCCAAAAT 3′
R: 5′ GGCTGTTGTCATACTTCTCATGG 3′
Cloning of the XLF Expression vector
The NHEJ1/XLF cDNA was cloned into the pcDNA4- HisMAX vector (Invitrogen) using the pcDNA4/HisMax TOPO TA Expression Kit (Thermo Fisher Scientific) per manufacturer’s instructions. Correct orientation of XLF cDNA was confirmed by Sanger sequencing.
Primers Used to Clone NHEJ1/XLF cDNA: F: 5′ATGGAAGAACTGGAGCAAGGCCTG 3′
R: 5′ TTAACTGAAGAGACCCCTTGGCTTC 3′
Cloning of the NHEJ1/XLF promoter reporter construct
A 1.62 kb region of the NHEJ1/XLF promoter, containing 1.5 kb upstream and 120 bp downstream of the NHEJ1/XLF transcription start site, was amplified from human genomic DNA using Platinum Pfx DNA Polymerase (Thermo Scientific) according to the manufacturer’s protocol. The primers used were: 5′-GATTCCTGGTAAGTTGAGGCTAGGCCCTAGC-3′ (Forward) and 5′-CTCCTGCCCGGACTCGAACGCGATTCCAC-3′ (Reverse). The amplified product was cloned into the pCR2.1-TOPO vector using the TOPO TA Cloning Kit (Invitrogen) according to the manufacturer’s protocol. The cloned sequence was confirmed by Sanger sequencing. The NHEJ1/XLF promoter was then subcloned into the pGL3-Basic Vector (Promega) using KpnI and XhoI restriction enzyme sites contained in both plasmids. The XLF promoter sequence in the pGL3 luciferase reporter vector was again confirmed by Sanger sequencing. Promoter deletions were made using the Q5 site directed mutagenesis kit (NE Biolabs) per manufacturer’s protocol.
NHEJ1/XLF promoter luciferase reporter assay
24 hours prior to transfection 7×104 cells were seeded in 12-well plates. Cells were transfected with Lipofectamine 2000 (Life Technologies) using 1 μg of pGL3-XLF Promoter and 50 ng pCMV-RL as a transfection efficiency control. Transfections were carried out in triplicate. Luciferase activity analyzed using the Dual Luciferase Reporter Assay Kit (Promega). Promoter activity was calculated by dividing the firefly luciferase activity from the NHEJ1/XLF promoter reporter vector by the transfection efficiency Renilla luciferase signal and multiplying the ratio by 1000.
Chromatin Immunoprecipitation-quantitative PCR (ChIP-qPCR)
ChIP-qPCR assays were performed as described (30). Beads alone were used as a negative control and results are presented as percent input, using the 1% input DNA sample for normalization. ChIP-qPCR experiments were performed as previously described (30). Briefly, after immunoprecipitation with indicated antibodies, precipitated DNA was combined with primers and Power SYBR Green PCR Master mix (Applied Biosystems/Life Technologies). Plates were spun down prior to analysis. The Mx3000p real time PCR system (Stratagene) was used to monitor fluorescence intensity in real-time to allow quantitative comparisons. Ct values were normalized to input using the −ΔΔCt method. Beads alone were used as a negative control, and results are presented as percent input, using the 1% input DNA sample for normalization.
Antibodies used for ChIP assays
Acetyl-K9 H3 (07-352, Millipore), Trimethyl-K9 H3 (17-625, Millipore), Dimethyl-histone-H3 (Lys9) (07-441, Upstate Cell Signaling Solutions), Acetyl-Histone H3(Lys27) (07-360, Millipore), PTEN (#9559, Cell Signaling Technologies), PCAF (#3378, Cell Signaling Technologies), CBP (#7389, Cell Signaling Technologies)
Primers used for ChIP-qPCR
NHEJ1/XLF Promoter: F: 5′ GCCTCGCCCGCTATTCTTTCCACTCG 3′
R: 5′ CTGCCCGGACTCGAACGCGATTCCAC 3′
Primers for FANCD2 promoter (31) and RAD51 (32) have been previously described.
Co-immunoprecipitation assays
Antibodies used were PCAF (#3378, Cell Signaling Technologies) and CBP (#7389, Cell Signaling Technologies) for IP and for blotting. Mouse anti-PTEN (sc-7974, Santa Cruz Biotechnology) was used to immunoprecipitate PTEN and for blotting after IP with PCAF and CBP antibodies. Rabbit anti-PTEN (#9552, Cell Signaling Technologies) was used to blot anti-PTEN immunoprecipitates. Cells were lysed with RIPA buffer and incubated at 4° C with primary antibodies. Immunoprecipitation assays were performed in the presence of genomic DNA.
Results
PTEN loss is associated with NHEJ deficiency in patient derived melanomas
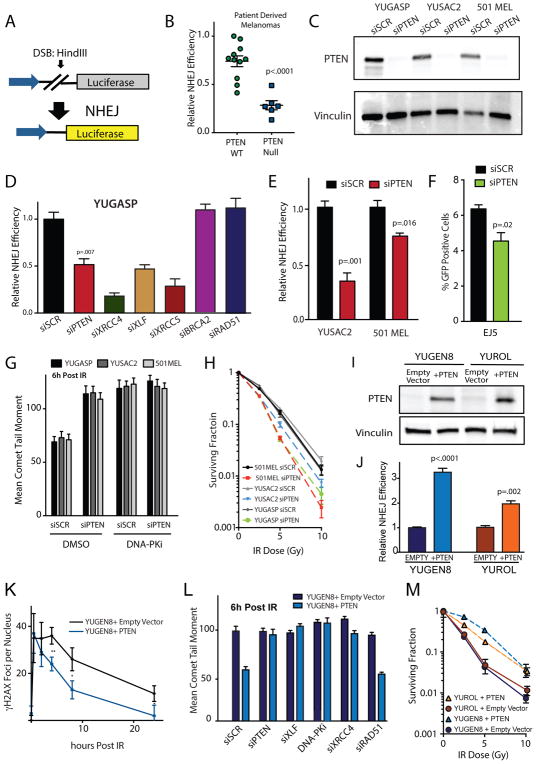
Though melanomas are traditionally a radioresistant cancer type, recent evidence indicates variability in in radiosensitivity of patient derived melanoma cultures (22). We hypothesized this variability may reflect novel mechanisms regulating the efficiency of the NHEJ pathway. We subsequently examined the NHEJ efficiencies of low passage patient-derived melanoma cultures and melanoma cell lines with key clinical characteristics shown in Supplementary Figure S1A using a host-cell reactivation reporter assay. The NHEJ host-cell reactivation assay (Figure 1A) was previously described (28, 33, 34) and reports as expected in matched pair cell lines (Supplementary Figure S1B). Specifically, the XRS6 cell line, which is deficient in the NHEJ factor Ku80, shows very low levels of NHEJ, but NHEJ activity is restored upon Ku80 complementation. In contrast, BRCA2 homozygous knockout in the DLD1 cells does not impair reactivation of the NHEJ reporter (Supplementary Figure S1B). Combining functional NHEJ results with analysis of whole exome sequencing data and gene expression data available for these melanomas (23), we found a strong correlation between PTEN status and NHEJ activity, as melanomas lacking PTEN expression [as previously documented in (23, 25) and as confirmed by western blot (Supplementary Figure S1C)] consistently showed lower levels of NHEJ (Figure 1B). No correlation with HR pathway activity and PTEN was found in the primary melanoma patient sample cohort using an HDR host cell reactivation assay (28, 35) (Supplementary Figure S1D).
Figure 1. PTEN loss suppresses NHEJ and PTEN complementation rescues the NHEJ defect in PTEN-null melanomas.
(A) Schematic representation of the NHEJ luciferase based reporter assay. (B) Relative NHEJ efficiency of short-term patient derived melanoma cultures. (C) Western blot analysis of PTEN expression in YUGASP, YUSAC2 and 501MEL cells 96 h after transfection of siRNA targeting PTEN. (D) Relative NHEJ efficiency of short-term patient derived melanoma culture YUGASP after siRNA suppression of PTEN, XRCC4, XLF, XRCC5, BRCA2, and RAD51. (E) Relative NHEJ efficiency of YUSAC2 and 501 melanoma cell lines after PTEN suppression by siRNA compared to scrambled control siRNA. (F) EJ5 end-joining reporter assay in U2OS cells performed after PTEN suppression by siRNA compared to scrambled control siRNA. (G) Neutral comet assay performed 6 h after IR in YUGASP, YUSAC2 and 501MEL cells 96 h transfection with siPTEN and 24 h after exposure to 10 μM DNA-PKi NU7441 or DMSO control. (H) Radiation survival with or without PTEN knockdown in YUGASP, YUSAC2 and 501MEL melanoma cells using siPTEN compared to siSCR. (I) Western blot analysis of PTEN expression in YUGEN8 and YUROL cultures complemented with either pBABE-PURO PTEN or pBABE-PURO empty vector. Vinculin is used as a loading control. (J) NHEJ reporter assay performed in YUGEN8 and YUROL with and without PTEN complementation. Reporter activity is normalized to empty vector control. (K) Quantification of γH2AX foci per nucleus 0 through 24 h post 5 Gy IR in YUGEN8 cultures complemented with either pBABE-PURO PTEN or pBABE-PURO empty vector. (L) Quantification of neutral comet assay performed in 6 h after 5 Gy IR in YUGEN8 cells complemented with either pBABE-PURO PTEN or pBABE-PURO empty vector, 96 h after knockdown of indicated factors with siRNA or 24 h after 10 μM DNA-PKi, NU7441. (M) Radiation survival, quantified by colony formation, of clonal PTEN complemented cell lines compared to empty vector control for YUGEN8 and YUROL. For all panels, error bars represent SEM (n=3) and statistical analysis by t-test.
PTEN suppression compromises NHEJ activity, while PTEN complementation rescues NHEJ
To further investigate the correlation of PTEN in patient-derived melanomas with NHEJ efficiency, we knocked down PTEN in a PTEN wild-type, short-term patient-derived melanoma culture, YUGASP (Figure 1C) in parallel with knockdown of other DSB repair factors: XRCC4, XLF, and XRCC5 (Ku80) in the NHEJ pathway and BRCA2 and RAD51 in the HDR pathway (Supplementary Figure S1E). PTEN knockdown as well as XRCC4, XRCC5, and XLF knockdown yielded significant decreases in NHEJ capacity (Figure 1D). We also suppressed PTEN with siRNA in the long-term cultured melanoma-derived cell lines, YUSAC2 and 501MEL (Figure 1C) and observed a decrease in NHEJ activity (Figure 1E), whereas siPTEN transfection into the PTEN null YUGEN8 cells had no effect on the already low NHEJ capacity of these cells (Supplementary Figure S1F). Additionally, knockdown of PTEN in U2OS cells (an osteosarcoma-derived line) with the EJ5 chromosomally integrated NHEJ reporter, an established benchmark assay in the field (36), showed a decrease in end joining activity (Figure 1F). PTEN knockdown impaired the repair of DSBs at 6 h post 5 Gy IR in the YUGASP, YUSAC2 and 501MEL cells, a time point indicative of NHEJ activity (Figure 1G). However, in the presence of the DNA-PK inhibitor, NU7441, siRNA knockdown of PTEN had no additional effect on DSB resolution detected by the neutral comet assay as compared to control siRNA, suggesting an epistatic relationship of PTEN knockdown with DNA-PK inhibition (Figures 1G), and placing PTEN function in the DNA-PK-mediated canonical NHEJ pathway (37, 38). Consistent with a decrease in NHEJ efficiency, we observed a relative radiosensitization of the otherwise PTEN wild-type YUGASP, YUSAC and 501MEL cells upon PTEN knockdown (Figure 1H).
To further investigate the relationship between PTEN and NHEJ, we complemented two PTEN null melanoma cultures, YUGEN8 and YUROL, with expression of wild-type PTEN cDNA using the pBABE-PURO retroviral gene delivery system. The transduced YUGEN8 and YUROL cell populations showed robust PTEN expression (Figure 1I) and exhibited marked increases in NHEJ efficiency (Figure 1J) but no effect on HDR in these cells (Supplementary Figure S2A). Consistent with the impact on NHEJ in the reporter assay, PTEN complementation also provided an increase in the resolution in γH2AX and p53BP1 foci post IR (Figure 1K and Supplementary Figure S2B and S2C) and a decrease in the persistence of DSBs 6 h post IR (Figure 1L), at time points indicative of NHEJ activity (32).
In addition, PTEN’s ability to increase the efficiency of DSB resolution at 6 after IR, measured by the comet assay, was abolished by siRNA knockdown of the ectopic PTEN (Figures 1L, S2D and S2E). To further investigate PTEN’s role in DSB repair, we conducted epistasis experiments in which we suppressed core components of the NHEJ and HDR pathways and measured persistence of DSBs at 6 h and 24 h after IR. The knockdown of the NHEJ components XLF and XRCC4 (Figure S2D), as well as chemical inhibition of DNA-PK resulted in abrogation of PTEN’s ability to promote DSB repair after IR in the comet assay (Figure 1L and Supplementary Figure S2E), suggesting that PTEN acts in the same pathway as XLF, XRCC4 and DNA-PK, the canonical end joining pathway. Rad51 knockdown showed an additive effect independent of PTEN status in reducing DSB repair measured by mean comet tail moment at 24 h post IR (Supplementary Figure S2E). In keeping with the NHEJ defect upon PTEN loss, we further observed, by clonogenic survival assay, that PTEN complementation provides a radio-protective effect on cell survival after IR relative to empty vector controls (Figure 1M). Similar to the melanoma data, doxycycline induced PTEN expression was able to increase the NHEJ efficiency in the PTEN-null U251 glioblastoma cell line (Supplementary Figure S2F).
PTEN regulates XLF levels
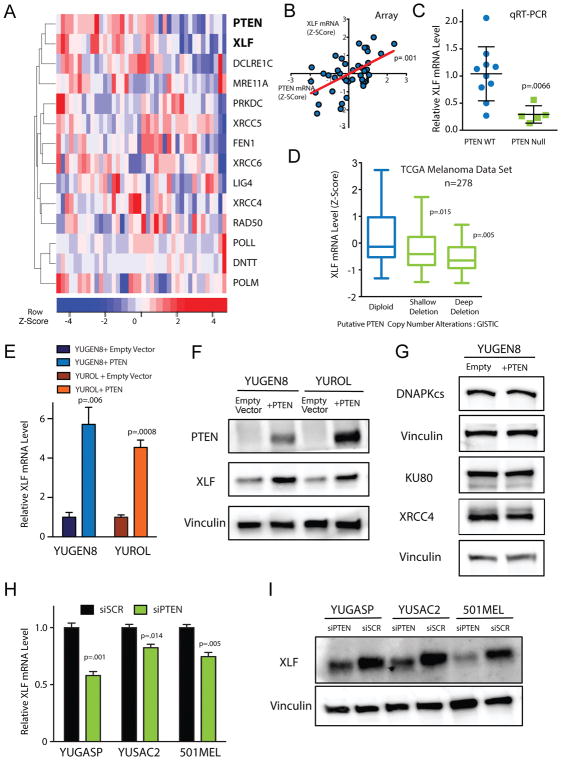
To investigate the mechanism by which PTEN promotes NHEJ activity, we analyzed microarray gene expression data from 40 patient-derived melanoma cultures in the Yale Melanoma Gene Expression Cohort (25). Unsupervised clustering of PTEN expression with genes coding for proteins in the NHEJ pathway revealed that the expression pattern of XLF (encoded by the NHEJ1 gene) clustered most closely to that of PTEN (Figure 2A). XLF is a core component of the NHEJ machinery where it participates in DNA end bridging and ligation through interaction and complex formation with XRCC4 and DNA ligase IV(5). Cells lacking XLF have been found to be profoundly impaired in DNA DSB repair and radiosensitive (39, 40). In addition, mutations in this gene (which is also known as Cernunnos) are linked to a human syndrome of growth retardation, microcephaly, immunodeficiency and cellular sensitivity to IR (6). Further analysis revealed that PTEN mRNA levels and XLF mRNA levels are highly and positively correlated (Figure 2B). In contrast, PTEN expression was not as highly correlated with any other NHEJ or HDR gene (nor with GAPDH as a control); data sets showing lack of correlation with RAD51, BRCA1, FANCD2 as well as GAPDH mRNA levels are presented as examples (Supplementary Figure S3A). We validated the microarray data using quantitative reverse transcription PCR (qRT-PCR) for XLF mRNA levels and confirmed that melanomas null for PTEN had low levels of XLF mRNA as compared to melanomas expressing PTEN (Figure 2C). Further to this trend, publicly available RNA-sequencing data from the Cancer Genome Atlas Network (TCGA) for cutaneous melanomas show a strong correlation between PTEN status and XLF mRNA levels (Figure 2D) (7). Additionally, publicly available microarray expression data from glioblastomas (Supplementary Figure S3B) (11) and RNA-sequencing data from prostate adenocarcinomas (Supplementary Figure S3C) (10) and uterine carcinomas (Supplementary Figure S3D) (9), three additional cancer types with high incidences of PTEN alteration, show the same correlation: tumors with loss of PTEN have low levels of XLF mRNA.
Figure 2. PTEN status is correlated with XLF expression.
(A) Unsupervised clustering of PTEN expression with expression of NHEJ genes in the Yale Melanoma Gene Expression Cohort. (B) Scatter plot of PTEN expression vs. XLF expression in the melanoma cohort (n=40). Two-tailed p-value was determined from correlation regression analysis. (C) Relative XLF mRNA levels across melanoma samples assayed by qRT-PCR. Error bars represent SEM of samples analyzed in triplicate. (D) Analysis of XLF mRNA levels as a function of PTEN copy number alteration in TCGA cutaneous melanoma RNA and DNA sequencing data (n=278). (E) Relative XLF mRNA levels in YUGEN8 and YUROL cells complemented with PTEN or empty vector control. mRNA levels were normalized to GAPDH mRNA and presented relative to empty vector control. Error bars represent SEM (n=3). (F) Western blot for XLF protein expression in YUGEN8 and YUROL cells complemented with PTEN or empty vector controls. Vinculin is used as a loading control. (G) Western blot analysis of DNA-PKcs, Ku80 and XRCC4 protein levels in YUGEN8 cells with or without PTEN complementation. Vinculin is used as a loading control. (H) Relative XLF mRNA levels in YUGASP, YUSAC2 and 501MEL cells, comparing PTEN siRNA knockdown to scrambled siRNA control. mRNA levels were normalized to GAPDH mRNA and presented relative to siSCR. Error bars indicate SEM (n=3). (I) Western blot of XLF expression comparing siPTEN with siSCR in YUGASP, YUSAC2 and 501MEL cells. Vinculin is used as a loading control. For (C), (D), (E) and (H) statistical analysis was by t-test.
To further investigate the correlation of PTEN with XLF expression, we modulated PTEN levels in the melanoma cultures and measured the impact on XLF expression. PTEN null melanomas complemented by PTEN cDNA expression showed marked increases in XLF mRNA and protein levels (Figure 2E and F) but did not show changes in the levels of the other core NHEJ proteins (Figure 2G). Conversely, PTEN knockdown in PTEN wild-type YUGASP, YUSAC2 and 501MEL cultures caused decreases in XLF mRNA and protein levels (Figures 2H and 2I). We also observed increases in XLF mRNA levels in U251 cells as a function of PTEN expression (Supplementary Figure S3E). PTEN suppression by siRNA in melanocytes induced a decrease in XLF mRNA and protein levels (Supplementary Figure 3F, G and H). PTEN knockout in HCT116 cells did not show a decrease in XLF mRNA levels (Supplementary Figure 3I).
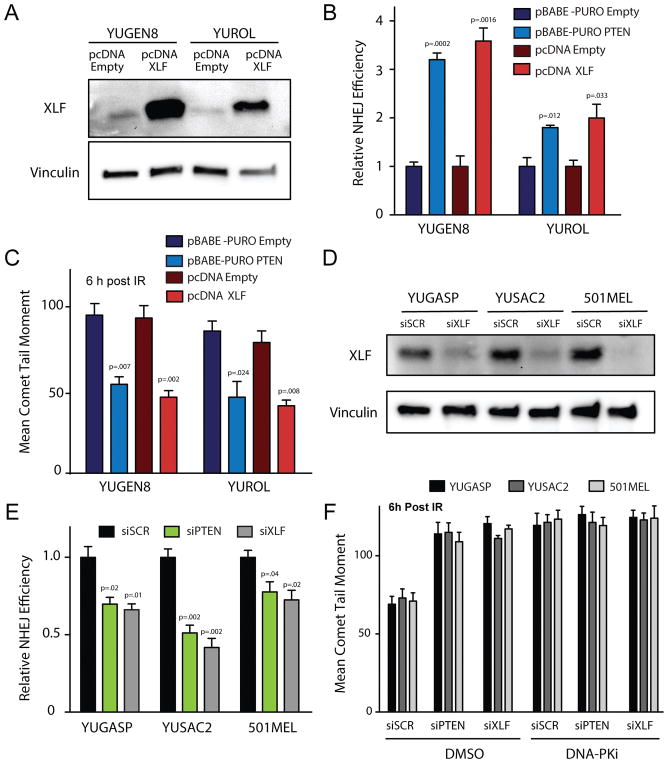
Restoration of XLF expression rescues NHEJ activity in PTEN null melanomas, while XLF knockdown in PTEN wild-type melanomas compromises NHEJ
To further test whether low XLF expression accounts for the diminished NHEJ activity in the PTEN null melanomas, we expressed XLF using an exogenous cDNA expression vector in the PTEN null, NHEJ-deficient melanomas (Figure 3A). We found that forced expression of XLF in these cells consistently boosted NHEJ activity to levels that were similar in magnitude to the effect of PTEN complementation in these cells (Figure 3B). Moreover, forced expression of XLF also yielded reduction of comet tail moments in these melanomas 6 h post 5 Gy IR, indicating functional increases in DSB repair activity by NHEJ, again similar to the impact of PTEN complementation (Figure 3C). On the other hand, XLF suppression using siRNA knockdown in PTEN wild-type cells (YUGASP, YUSAC2 and 501MEL) produced decreases in NHEJ efficiency, to an extent similar to the effect of PTEN knockdown (Figures 3D and 3E). We also used the neutral comet assay to measure the impact of XLF knockdown on DSB repair, and we observed an increase in comet tail moment indicative of reduced DSB repair (Figure 3F), again similar to the results of PTEN knockdown. In the presence of the DNA-PK inhibitor, NU7441, siRNA knockdown of XLF had no additional effect on DSB repair in the comet assay as compared to control siRNA, consistent with the known epistatic relationship of XLF with DNA-PK and similar to what was observed for DNA-PK inhibition combined with PTEN knockdown (as shown in Figure 1G and reproduced for comparison in Fig. 3F).
Figure 3. XLF overexpression rescues NHEJ deficiency in PTEN-null melanomas and phenocopies PTEN complementation while XLF suppression phenocopies PTEN suppression with respect to NHEJ activity.
(A) Western blot analysis of XLF expression 72 h after transfection of YUGEN8 and YUROL with wild-type XLF expression vector (pcDNA XLF) or empty vector control (pcDNA Empty). Vinculin is used as a loading control. (B) NHEJ reporter assay comparing PTEN-null YUGEN8 and YUROL cells with or without PTEN complementation versus with or without XLF cDNA expression. Reporter activity is normalized to the empty vector controls. (C) Quantification of neutral comet assay analysis to measure persisting DNA DSBs at 6 h after 5 Gy IR in YUGEN8 and YUROL cells with or without PTEN complementation versus with or without XLF cDNA expression. (D) Western blot analysis confirming XLF knockdown by siRNA 72 h after siRNA transfection. Vinculin is used as a loading control. (E) NHEJ reporter assay analysis in YUGASP, YUSAC2, and 501MEL cells after siRNA knockdown of PTEN (siPTEN), XLF (siXLF) or scrambled control siRNA (siSCR). Reporter activity is normalized to siSCR. (F) Quantification of neutral comet assay analysis of persisting DNA DSBs 6 h after 5 Gy IR in YUGASP, YUSAC2 and 501MEL cell lines transfected with siPTEN, siXLF or siSCR and treated with either DMSO or 10 μM DNA-PK inhibitor (NU7441) (>100 cells analyzed/replicate). In panels (B) and (E) error bars represent SEM (n=3). In (C) and F error bars represent SEM (n=3) with >100 cells analyzed, and statistical analysis by t-test.
PTEN recruits the transcription co-activators and histone acetyltransferases, PCAF and CBP, to the XLF promoter
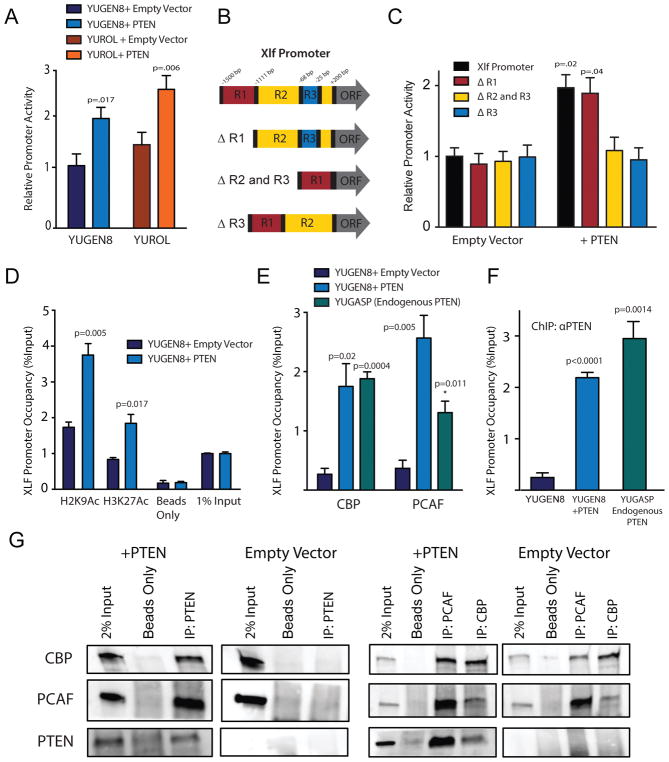
To elucidate a mechanistic basis of XLF regulation by PTEN, we first tested for an effect of PTEN on transcription from a construct containing the XLF promoter driving a luciferase reporter gene. We found that PTEN complementation in the PTEN null melanomas increases transcription from the NHEJ1/XLF promoter (Figure 4A) but not from the FANCD2 or SV40 promoters (Supplementary Figure S4A). To elucidate the specific region of the NHEJ1/XLF promoter under regulation by PTEN, we performed deletion analysis of the NHEJ1/XLF promoter reporter construct revealing that a 43 bp region from 25 to 68 bp (termed R3) upstream of the NHEJ1/XLF transcription start site was necessary for the PTEN-dependent increase in promoter activity (Figures 4B and 4C). Next, we examined the epigenetic status of this region in the endogenous NHEJ1/XLF promoter as a function of PTEN status by chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) with an amplicon centered upon the endogenous R3 sequence necessary for PTEN-dependent transcription from the reporter construct. Specifically, we performed ChIP-qPCR to quantify levels of Histone 3 Lysine 9 (H3K9) acetylation and Histone 3 Lysine 27 (H3K27) acetylation at this region of the NHEJ1/XLF promoter in the clonal matched pair YUGEN8 cell lines complemented with either pBABE-PURO Empty Vector or pBABE-PURO PTEN. We observed a significant PTEN-dependent increase in the activating H3K9 acetylation and H3K27 acetylation marks at the NHEJ1/XLF promoter (Figure 4D).
Figure 4. Induction of XLF expression by epigenetic activation of the XLF Promoter by the histone acetyltransferases, PCAF and CBP, as a function of PTEN.
(A) XLF promoter-luciferase expression assay in PTEN-null YUGEN8 and YUROL cells with or without PTEN cDNA complementation. (B) Schematic of deletion analysis of the XLF promoter reporter construct. (C) Quantification of deletion analysis of XLF promoter reporter activity. Data is normalized to full-length promoter activity in the YUGEN8 + Empty Vector sample. (D) Epigenetic status of the indicated histone marks at the XLF promoter in clonal YUGEN8 cell lines with or without PTEN complementation as determined by ChIP-qPCR. (E) XLF promoter occupancy by PCAF and CBP in clonal YUGEN8 cells as a function of PTEN expression, and in YUGASP cells expressing endogenous wild-type PTEN as determined by ChIP-qPCR. (F) XLF promoter occupancy by PTEN in clonal YUGEN8 cells with or without PTEN complementation and in YUGASP cells expressing endogenous PTEN. (G) Co-IP western blot analyses in YUGEN8 cells with or without PTEN complementation. IP was performed with antibodies to PTEN, CBP or PCAF, as indicated, and followed by western blot for all three factors. For (A–F) error bars represent SEM (n=3), and statistical analysis by t-test.
We hypothesized that the observed increases in H3K9 and H3K27 acetylation at the NHEJ1/XLF promoter could reflect the action of specific histone acetyltransferases, particularly PCAF and CBP (41–44), and so we probed for their potential interaction with the XLF promoter. We performed ChIP-qPCR with antibodies to either PCAF or CBP in the PTEN null versus PTEN-complemented YUGEN8 cells, revealing PTEN-dependent occupancy of the NHEJ1/XLF promoter by both PCAF and CBP (Figure 4E) as well as the active, acetylated form of CBP (Supplementary Figure S4B). Additionally, we confirmed that NHEJ1/XLF promoter occupancy by PCAF and CBP similarly occurs in melanoma cells that endogenously express wild-type PTEN (YUGASP melanoma cells; Figure 4E) but not in colon cancer cells (Supplementary Figure S4C), a cancer type not typically associated with PTEN mutation or loss. PCAF and CBP have broad acetyltransferase activity and require adaptor molecules to guide them to loci for site-specific histone acetylation (43), and so we hypothesized that PTEN, itself, might recruit these factors to the NHEJ1/XLF promoter. To investigate this, we conducted ChIP assays with a PTEN-specific antibody, revealing that PTEN can be detected at the XLF promoter in YUGEN8 cells complemented with PTEN (as well as in the YUGASP cells expressing endogenous wild-type PTEN), but not in the PTEN null YUGEN8 cells (Figure 4F). This PTEN interaction with the NHEJ1/XLF promoter appeared specific, as we observed no PTEN occupancy at either the RAD51 or FANCD2 promoters (genes in the HDR pathway) or in the promoter for the DCLRE1C gene (encoding the NHEJ factor, Artemis) (Figures S4D, S4E and S4F), nor did we detect any PTEN-dependent changes in PCAF or CBP occupancy or in the H3K9 or H3K27 acetylation marks at these promoters (Figures S4D, S4E and S4F).
Next, we tested for potential interactions between PCAF and/or CBP and PTEN that could provide the mechanistic link in the recruitment of these activating factors to the NHEJ1/XLF promoter by PTEN. In fact, PTEN had previously been shown to interact with PCAF in human 293T cells, in which case PCAF was reported to acetylate PTEN at lysines 125 and 128 and thereby inactivate its phosphatase activity (45). To test for physical interaction of PTEN with PCAF and/or CBP in the melanomas, we assayed for co-immunoprecipitation (co-IP) of these factors. We observed that IP with an antibody to PTEN pulls down both PCAF and CBP in lysates from PTEN-complemented YUGEN8 cells (Figure 4G), whereas there was no IP of these factors from lysates of the empty vector (and therefore still PTEN null) YUGEN8 cells (Figure 4G). In reverse, IP with an antibody to PCAF pulls down PTEN, as does an antibody to CBP, but again only in the PTEN-complemented and not in the PTEN null cells (Figure 4G). PCAF and CBP are also seen to co-IP with each other regardless of PTEN status (Figure 4G). In the YUGASP cells, IP of PTEN pulls down PCAF and CBP (Supplementary Figure S4G), and IP with antibodies to either PCAF or CBP pulls down PTEN (Supplementary Figure S4G), confirming that these interactions occur with endogenous wild-type PTEN. Taken together, these results demonstrate physical interaction among PTEN, PCAF and CBP and suggest a novel functional role for such interaction: to specifically direct epigenetic modification to the NHEJ1/XLF promoter.
PTEN acetylation at K128 is necessary to promote NHEJ through XLF induction in a pathway that is independent of PTEN phosphatase activity
Previously, PTEN had been shown to physically interact with PCAF resulting in acetylation of PTEN at the lysine 128 (K128) residue, thereby inactivating the phosphatase function of PTEN (45). We hypothesized this residue may have additional biological relevance with respect to regulation of nuclear PTEN function, particularly its role in regulating NHEJ. Notably, PTEN K128 has been found to be mutated in melanomas (46), breast (47), lung squamous cell (48), and uterine carcinomas (9), further suggesting functional importance. To test the role of PTEN acetylation at residue K128, we complemented YUGEN8 cells with the PTEN K128R variant in which the lysine at position 128 is substituted by an arginine (K128R), thereby blocking acetylation at this site. After complementation of YUGEN8 cells with PTEN K128R we do not observe a rescue of NHEJ activity (Figure 5A). Notably, PTEN K128R is still capable of nuclear localization (Supplementary Figure S5A) as well as suppression of AKT phosphorylation on S473 (Supplementary Figure S5B), suggesting that this is a separation of function mutation. Since acetylation of K128 has been reported to cause inactivation of the PTEN phosphatase activity, we next tested the dependence on PTEN’s phosphatase activity on stimulation of NHEJ using the biochemically well characterized, and well-studied phosphatase-dead PTEN C124S and PTEN G129E variants (49, 50). We found that, in contrast to the K128R variant, both PTEN C124S and G129E were able to rescue the NHEJ defect in the PTEN null YUGEN8 cells (Figure 5A). Accordingly, the C124S and G129E variants but not the K128R variant were able to induce an increase in XLF mRNA (Figure 5B) and protein levels (Figure 5C). To further evaluate the end joining defect associated with the PTEN K128R variant, we performed the neutral comet assay after 5 Gy IR (Figures 5D and E), and we observed a decrease in persisting DNA DSBs as compared to empty vector control in YUGEN8 cells overexpressing wild type PTEN, PTEN C124S and PTEN G129E but not those expressing PTEN K128R. Together, these results demonstrate that PTEN regulation of NHEJ does not depend on its phosphatase activity but does depend on K128 as a putative target for regulatory acetylation by PCAF.
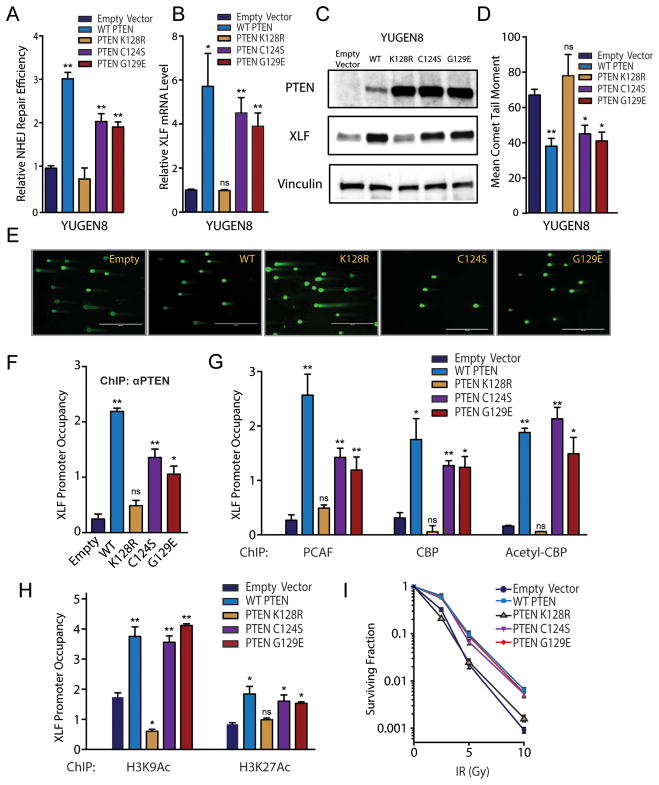
Figure 5. PTEN promotion of NHEJ is dependent on PTEN acetylation site K128 but is independent of PTEN phosphatase activity.
(A) NHEJ reporter assay in YUGEN8 cells complemented with empty vector control, wild type PTEN, PTEN K128R, PTEN C124S, or PTEN G129E, as indicated. Reporter activity is normalized to the YUGEN8 + empty vector control sample. (B) XLF mRNA levels in YUGEN8 cells complemented as indicated. mRNA levels were normalized to GAPDH mRNA and presented relative to empty vector control. (C) Western blot analysis of PTEN and XLF levels in YUGEN8 cells complemented as indicated. Vinculin is used as a loading control. (D) Quantification and (E) representative images of neutral comet assays performed 24 h after 5 Gy IR in YUGEN8 cells complemented as indicated. (F) PTEN occupancy of the XLF promoter assayed by ChIP-qPCR in YUGEN8 cells complemented as indicated. (G) PCAF, CBP, and acetyl-CBP occupancy of the XLF promoter in YUGEN8 cells complemented as indicated. (H) H3K9 acetylation and H3K27 acetylation at the XLF promoter in YUGEN8 cells complemented as indicated. (I) Radiation survival curves quantified by clonogenic survival of pooled YUGEN8 cell lines complemented as indicated. For (A), (B) and (D), error bars represent SEM (n=3). (F), (G) and (H) PTEN mutants were analyzed in triplicate and error bars represent SEM. ChIP-qPCR data for YUGEN8 complemented with wild type PTEN or empty vector appear in Figure 6. For all panels statistical analysis by t-test (* = p<.05 and ** = p<.01).
To further probe the mechanistic importance of the PTEN K128 residue, we tested whether abrogation of this acetylation site disrupts PTEN’s ability to epigenetically modulate XLF expression. We found by ChIP-qPCR that the PTEN K128R variant shows substantially less occupancy of the NHEJ1/XLF promoter as compared to wild-type PTEN or to the phosphatase variants, PTEN C124S and PTEN G129E (Figure 5F). In PTEN null YUGEN8 cells, expression of PTEN K128R fails to stimulate NHEJ1/XLF promoter occupancy of PCAF, CBP or active acetyl-CBP (Figure 5G) and does not yield any increases in the active chromatin modifications of H3K9 acetylation and H3K27 acetylation at the NHEJ1/XLF promoter (Figure 5H). In contrast, the phosphatase dead PTEN C124S and PTEN G129E, like wild type PTEN, both induce PCAF, CBP and acetyl-CBP occupancy of the NHEJ1/XLF promoter (Figure 5G) and thereby stimulate H3K9 acetylation and H3K27 acetylation (Figure 5H). These results further indicate a separation of function between PTEN mutants, demonstrating that PTEN occupancy of the NHEJ1/XLF promoter and subsequent recruitment of the PCAF and CBP histone acetyltransferases is dependent on PTEN acetylation at lysine 128 but is independent of PTEN’s phosphatase activity, establishing a mechanistic basis for the PTEN regulation of NHEJ.
Next, we performed clonogenic survival assays on pooled populations of YUGEN8 cells transduced with pBABE-PURO empty vector control, pBABE-PURO PTEN, pBABE-PURO PTEN K128R, pBABE-PURO PTEN C124S and pBABE-PURO PTEN G129E. We observed substantial radioprotection conferred by expression of wild-type PTEN and by the phosphatase inactivating variants C124S and G129E, but not by the K128R variant (compared to the empty vector) (Figure 5I), consistent with the data that wild-type PTEN as well as the C124S and G129E variants can complement the NHEJ defect of PTEN null cells by induction of XLF expression whereas the K128R variant cannot promote end joining.
Discussion
In the work reported here, we have identified PTEN as a regulator of the NHEJ pathway through a novel mechanism of epigenetic induction of XLF expression that is dependent upon PTEN K128 but is independent of PTEN phosphatase activity. By interrogating functional DNA repair capacity in a panel of patient-derived human melanomas for which whole exome sequencing and gene expression data are available (23–25), we discovered an association between PTEN loss and NHEJ deficiency. We confirmed a causal relationship between PTEN and NHEJ by showing that complementation of PTEN null melanomas with PTEN increases NHEJ activity, and furthermore this increase in NHEJ can be abolished by knockdown of the ectopic PTEN with siRNA. Moreover, siRNA suppression of PTEN in PTEN wild-type melanomas reduces NHEJ. We further established an epistatic relationship between PTEN depletion and inhibition of DNA-PK as well as with knockdown of core canonical NHEJ factors in the repair of IR-induced DNA DSBs, placing this function of PTEN in the DNA-PK dependent canonical NHEJ pathway (38). Through analysis of the Yale Melanoma Cohort and of publically available human cancer genomics data, we detected a strong correlation of PTEN status with XLF mRNA levels in melanoma, prostate cancer, uterine cancer and glioblastoma. Mechanistically, we demonstrated transcriptional regulation of the NHEJ factor XLF by PTEN, and we determined that this regulation mediates the effect of PTEN on NHEJ activity. We demonstrated that XLF knockdown or overexpression phenocopies that of PTEN, while knockdown of XLF in the context of PTEN complementation abolishes PTEN’s ability to stimulate NHEJ. Further to mechanism, we found that PTEN occupies the NHEJ1/XLF promoter and physically interacts with the transcriptional co-activators and histone acetyltransferases, PCAF and CBP. These factors were also found by ChIP to occupy the NHEJ1/XLF promoter in a PTEN-dependent manner, suggesting their recruitment to the promoter by PTEN. We determined PTEN residue K128 mediates the PTEN-dependent epigenetic activation of XLF independent of PTEN phosphatase activity. Taken together, these results reveal a novel, K128-dependent nuclear function of PTEN: specific epigenetic activation of the XLF promoter to promote efficient NHEJ activity.
PTEN is the second most frequently mutated or lost tumor suppressor gene in human cancers, with PTEN alterations found in approximately 8% of all human cancers (51, 52), notably including melanomas (7) and glioblastomas (11). We observe a PTEN-dependent downregulation of XLF in melanoma cultures, melanocytes, and U251 glioma cells. However, after PTEN knockout in HCT116 colorectal cancer cells we do not observe PTEN-dependent decrease in XLF levels, indicating that this phenomenon might not be present in cancer types that are not associated with PTEN loss.
Beyond the identification of PTEN regulation of the NHEJ pathway, our results establish a novel molecular mechanism for this regulation: PTEN interaction with PCAF and CBP to epigenetically activate the NHEJ1/XLF promoter. We observed PTEN-dependent increases in active histone marks at the NHEJ1/XLF promoter, characteristic of PCAF and CBP activity. PCAF and CBP both have broad acetyltransferase function and can transfer an acetyl group to a wide range of substrates; for CBP and PCAF to act upon histones at a specific promoter, an activator protein or complex cab be needed to target them there. Our results indicate that PTEN may function as such an activator for the NHEJ1/XLF gene, as PCAF, CBP and PTEN occupy the NHEJ1/XLF promoter in a PTEN-dependent manner. Further, reciprocal co-IP results show that PTEN physically interacts with PCAF and CBP, again pointing to a mechanism whereby PTEN recruits PCAF and CBP to the XLF promoter. Interestingly, prior work has shown that PCAF can acetylate nuclear PTEN upon physical interaction of the two proteins, thereby inactivating PTEN’s phosphatase activity (45) consistent with a report that nuclear PIP3 levels are unchanged as a function of PTEN expression (53). Therefore, we hypothesized that the phosphatase activity of PTEN would likely be dispensable for a nuclear function of epigenetically inducing XLF expression. By complementing PTEN null melanomas with PTEN C124S and G129E phosphatase mutants in comparison to PTEN K128R we determined PTEN stimulation of the NHEJ pathway is independent of PTEN phosphatase activity but is dependent on residue K128, a known target for regulatory acetylation. This finding may explain some of the discrepancies among studies evaluating the role of PTEN in DNA repair, as we find that different mutations in PTEN do not all act alike with respect to NHEJ.
Interestingly, a recent report suggests that AKT phosphorylates XLF, targeting it for degradation (54). In combination, our work and this reported role of AKT in post-translational modification of XLF together indicate that XLF is the target of multilevel regulation: post-translational regulation by the PTEN/AKT/PI3K axis and transcriptional regulation by phosphatase independent actions of nuclear PTEN. These complementary findings establish XLF as a key regulatory node for the NHEJ pathway, consistent with XLF deficiency leading to NHEJ defects and radiosensitization (39, 40) and with germline XLF gene mutations causes a human syndrome of developmental abnormalities and immunodeficiency (6).
Overall, using a panel of primary patient-derived melanoma cultures, along with manipulation of PTEN status, we have been able to evaluate the impact of PTEN on several DNA repair pathways, and we have established a novel nuclear function for PTEN: promotion of NHEJ through epigenetic regulation of XLF. These results highlight PTEN loss as a potential biomarker for NHEJ deficiency in human melanomas. These results may also provide the basis to devise targeted synthetic lethal strategies for melanoma treatment and may help to guide the design of new personalized cancer therapies.
Supplementary Material
Implications.
PTEN is the second most frequently lost tumor suppressor gene. Here it is demonstrated that PTEN has a direct and novel regulatory role in non-homologous end joining, a key DNA repair pathway in response to radiation and chemotherapy.
Acknowledgments
We thank Ruth Halaban, Denise Hegan, and Yanfeng Liu for their help. This work was supported by R35CA197574 and R01ES005775 to PMG and by the Specimen Resource Core of the Yale SPORE in Skin Cancer (P50CA121974 to Ruth Halaban).
Footnotes
Conflict of Interest Statement:
We have no conflicts of interest.
References
- 1.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 2.Fattah FJ, Lichter NF, Fattah KR, Oh S, Hendrickson EA. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc Natl Acad Sci USA. 2008;105:8703–8. doi: 10.1073/pnas.0712060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci USA. 2009;106:12430–5. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes & development. 1999;13:916–34. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 5.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–13. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Buck D, Malivert L, de Chasseval R, Barraud A, Fondanèche M-C, Sanal O, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–99. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Network CGA. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes & development. 2006;20:2149–82. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 9.Network CGAR. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grasso CS, Wu YM, Robinson DR, Cao X. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–52. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–70. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 14.Fraser M, Zhao H, Luoto KR, Lundin C, Coackley C, Chan N, et al. PTEN deletion in prostate cancer cells does not associate with loss of RAD51 function: implications for radiotherapy and chemotherapy. Clinical Cancer Research. 2012;18:1015–27. doi: 10.1158/1078-0432.CCR-11-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee C, Kim JS, Waldman T. PTEN gene targeting reveals a radiation-induced size checkpoint in human cancer cells. Cancer Research. 2004;64:6906–14. doi: 10.1158/0008-5472.CAN-04-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mereniuk TR, El Gendy M. Synthetic lethal targeting of PTEN-deficient cancer cells using selective disruption of polynucleotide kinase/phosphatase. Molecular Cancer therapeutics. 2013;12:2135–44. doi: 10.1158/1535-7163.MCT-12-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassi C, Ho J, Srikumar T, Dowling RJO, Gorrini C, Miller SJ, et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341:395–9. doi: 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta A, Yang Q, Pandita RK, Hunt CR, Xiang T. Cell cycle checkpoint defects contribute to genomic instability in PTEN deficient cells independent of DNA DSB repair. Cell Cycle. 2009;8:2189–210. doi: 10.4161/cc.8.14.8947. [DOI] [PubMed] [Google Scholar]
- 19.Choi BH, Chen Y, Dai W. Chromatin PTEN is involved in DNA damage response partly through regulating Rad52 sumoylation. Cell Cycle. 2013;12:3442–7. doi: 10.4161/cc.26465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Sun Z, Huang C, He J, Lamb KL, Kang X, Gu T. PTEN C-terminal deletion causes genomic instability and tumor development. Cell Reports. 2014;6:844–54. doi: 10.1016/j.celrep.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahbazian D, Bindra RS, Kluger HM, Glazer PM. Radiation sensitivity and sensitization in melanoma. Pigment cell & melanoma research. 2013:26. doi: 10.1111/pcmr.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krauthammer M, Kong Y, Bacchiocchi A, Evans P, Pornputtapong N, Wu C, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parisi F, Micsinai M, Strino F, Ariyan S, Narayan D, Bacchiocchi A, et al. Integrated analysis of tumor samples sheds light on tumor heterogeneity. Yale J Biol Med. 2012;85:347–61. [PMC free article] [PubMed] [Google Scholar]
- 26.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic acids research. 2000;28:2585–96. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitt RJ, Georgescu M-M, Pollak M. PTEN-induction in U251 glioma cells decreases the expression of insulin-like growth factor binding protein-2. Biochemical and biophysical research communications. 2005;336:1056–61. doi: 10.1016/j.bbrc.2005.08.229. [DOI] [PubMed] [Google Scholar]
- 28.Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai H, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Science translational medicine. 2017;9:eaal2463. doi: 10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scanlon SE, Glazer PM. Hypoxic stress facilitates acute activation and chronic downregulation of Fanconi anemia proteins. Molecular Cancer Research. 2014;12:1016–28. doi: 10.1158/1541-7786.MCR-13-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, Wajapeyee N, Turker MS, Glazer PM. Silencing of the DNA mismatch repair gene MLH1 induced by hypoxic stress in a pathway dependent on the histone demethylase LSD1. Cell reports. 2014;8:501–13. doi: 10.1016/j.celrep.2014.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoskins EE, Gunawardena RW, Habash KB, Wise-Draper TM, Jansen M, Knudsen ES, et al. Coordinate regulation of Fanconi anemia gene expression occurs through the Rb/E2F pathway. Oncogene. 2008;27:4798–808. doi: 10.1038/onc.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bindra R, Glazer P. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007;26:2048–57. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- 33.Zhong Q, Chen C-F, Chen P-L, Lee W-H. BRCA1 facilitates microhomology-mediated end joining of DNA double strand breaks. J Biol Chem. 2002;277:28641–7. doi: 10.1074/jbc.M200748200. [DOI] [PubMed] [Google Scholar]
- 34.Zhuang J, Zhang J, Willers H, Wang H, Chung JH, van Gent DC, et al. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res. 2006;66:1401–8. doi: 10.1158/0008-5472.CAN-05-3278. [DOI] [PubMed] [Google Scholar]
- 35.Bahal R, McNeer NA, Quijano E, Liu Y, Sulkowski P, Turchick A, et al. In vivo correction of anaemia in β-thalassemic mice by γPNA-mediated gene editing with nanoparticle delivery. Nature Communications. 2016:7. doi: 10.1038/ncomms13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. DNA Repair Protocols. 2012:379–91. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 37.Davidson D, Amrein L, Panasci L, Aloyz R. Small Molecules, Inhibitors of DNA-PK, Targeting DNA Repair, and Beyond. Front Pharmacol. 2013;4:5. doi: 10.3389/fphar.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Zeng ZC, Perrault AR, Cheng X, Qin W, Iliakis G. Genetic evidence for the involvement of DNA ligase IV in the DNA-PK-dependent pathway of non-homologous end joining in mammalian cells. Nucleic Acids Res. 2001;29:1653–60. doi: 10.1093/nar/29.8.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–13. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 40.Fattah FJ, Kweon J, Wang Y, Lee EH, Kan Y, Lichter N, et al. A role for XLF in DNA repair and recombination in human somatic cells. DNA repair. 2014;15:39–53. doi: 10.1016/j.dnarep.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641– 3. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 42.Jin Q, Yu L-R, Wang L, Zhang Z, Kasper LH, Lee J-E, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–62. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–24. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 44.Zhao X, Benveniste EN. Transcriptional activation of human matrix metalloproteinase-9 gene expression by multiple co-activators. J Mol Biol. 2008;383:945–56. doi: 10.1016/j.jmb.2008.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okumura K, Mendoza M, Bachoo RM, DePinho RA, Cavenee WK, Furnari FB. PCAF modulates PTEN activity. J Biol Chem. 2006;281:26562–8. doi: 10.1074/jbc.M605391200. [DOI] [PubMed] [Google Scholar]
- 46.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Network CGAR. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci USA. 1998;95:13513–8. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang XC, Piccini A, Myers MP, Van Aelst L, Tonks NK. Functional analysis of the protein phosphatase activity of PTEN. Biochemical Journal. 2012;444:457–64. doi: 10.1042/BJ20120098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao J, Aksoy BA, Dogrusoz U, Dresdner G. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6 doi: 10.1126/scisignal.2004088. pp.pl1-pl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindsay Y, McCoull D, Davidson L, Leslie NR. Localization of agonist-sensitive PtdIns (3, 4, 5) P3 reveals a nuclear pool that is insensitive to PTEN expression. Journal of cell science. 2006;119:5160–8. doi: 10.1242/jcs.000133. [DOI] [PubMed] [Google Scholar]
- 54.Liu P, Gan W, Guo C, Xie A, Gao D, Guo J, et al. Akt-Mediated Phosphorylation of XLF Impairs Non-Homologous End-Joining DNA Repair. Molecular cell. 2015;57:648–61. doi: 10.1016/j.molcel.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.