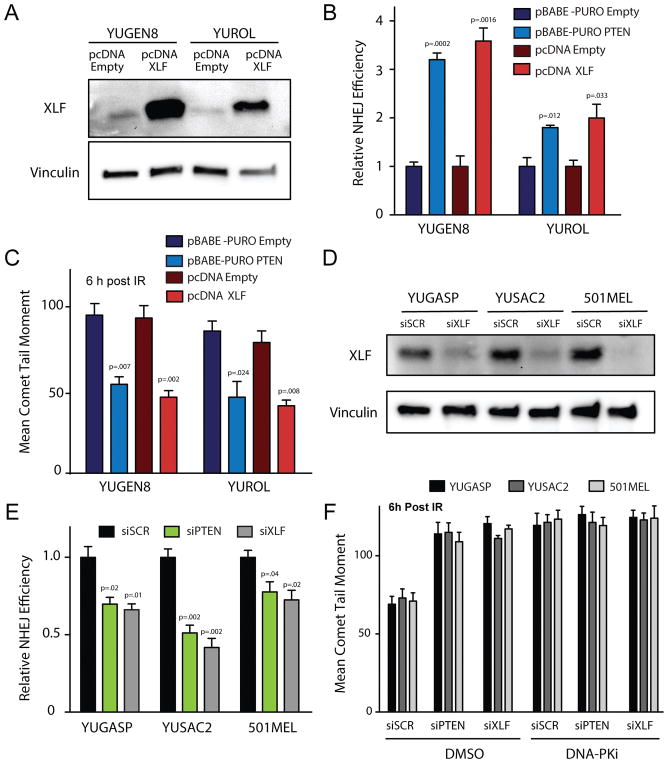
Figure 3. XLF overexpression rescues NHEJ deficiency in PTEN-null melanomas and phenocopies PTEN complementation while XLF suppression phenocopies PTEN suppression with respect to NHEJ activity.
(A) Western blot analysis of XLF expression 72 h after transfection of YUGEN8 and YUROL with wild-type XLF expression vector (pcDNA XLF) or empty vector control (pcDNA Empty). Vinculin is used as a loading control. (B) NHEJ reporter assay comparing PTEN-null YUGEN8 and YUROL cells with or without PTEN complementation versus with or without XLF cDNA expression. Reporter activity is normalized to the empty vector controls. (C) Quantification of neutral comet assay analysis to measure persisting DNA DSBs at 6 h after 5 Gy IR in YUGEN8 and YUROL cells with or without PTEN complementation versus with or without XLF cDNA expression. (D) Western blot analysis confirming XLF knockdown by siRNA 72 h after siRNA transfection. Vinculin is used as a loading control. (E) NHEJ reporter assay analysis in YUGASP, YUSAC2, and 501MEL cells after siRNA knockdown of PTEN (siPTEN), XLF (siXLF) or scrambled control siRNA (siSCR). Reporter activity is normalized to siSCR. (F) Quantification of neutral comet assay analysis of persisting DNA DSBs 6 h after 5 Gy IR in YUGASP, YUSAC2 and 501MEL cell lines transfected with siPTEN, siXLF or siSCR and treated with either DMSO or 10 μM DNA-PK inhibitor (NU7441) (>100 cells analyzed/replicate). In panels (B) and (E) error bars represent SEM (n=3). In (C) and F error bars represent SEM (n=3) with >100 cells analyzed, and statistical analysis by t-test.