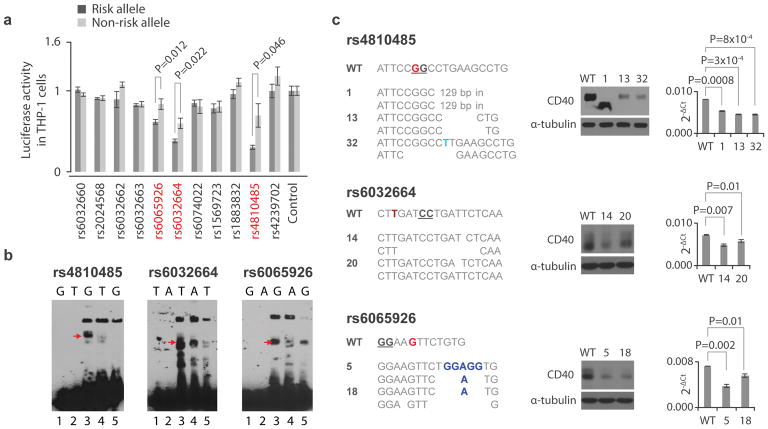
Figure 3. Validation of fSNPs rs4810485, rs6032664, rs6065926, rs1883832, and rs6074022 as CD40 fSNPs.
A. Reporter assay showing relative luciferase activity in human THP-1 cells between the risk (black) and non-risk (grey) alleles of 11 CD40 SNPs. Candidate fSNPs identified by SNP-seq highlighted in red (mean +/− SD, n=3 biological replicates, t-test with 2 tails without correction for multiple hypothesis testing). B. EMSA showing allele-specific gel shifting (arrows, n=3 independent biological replicates with similar results). rs4810485, G: risk/major allele, T: non-risk/minor allele; rs6032664, T: risk/major allele, A: non-risk/minor; rs6065926: G; risk/major, A: non-risk/minor; rs1883832: C: risk/major allele, T: non-risk/minor allele; and rs6074022, C: risk/major allele, T: non-risk/minor allele; Red arrow, allele-specific shift. Lane 1: risk allele probe only; lane 2: non-risk allele probe only; lane 3:risk allele probe with nuclear extract; lane 4: non-risk allele probe with nuclear extract; lane 5: risk allele probe with nuclear extract and excess unlabeled probe as cold competitor. C. CRISPR/Cas9 targeting rs4810485 (upper), rs6032664 (middle) and rs6065926 (lower) in the human B cell line BL2. Left: sequences showing mutations at the three SNP sites; Middle: Western blots (n=3 independent replicates with similar results); and Right: qPCR showing CD40 expression in all mutants (mean +/− SD, n=3 biological replicates, t-test with 2 tails). Red nucleotides (nts) represent the fSNPs. The genotype of WT BL2 cells at the three SNPs is homozygous. Blue nts reflect insertion; underlined nts indicate the NGG PAM for CRISPR/Cas9 targeting; in: insertion. WT: control for CRISPR/Cas9. Numbers indicate mutant clone designations.