Abstract
Background
Total nicotine equivalents (TNE), the sum of nicotine and metabolites in urine, is a valuable tool for evaluating nicotine exposure. Most methods for measuring TNE involve two-step enzymatic hydrolysis for indirect quantification of glucuronide metabolites. Here, we describe a rapid, low-cost direct liquid chromatography – tandem mass spectrometry (LCMS) assay.
Methods
In 139 smokers’ urine samples, Bland-Altman, correlation, and regression analyses were used to investigate differences in quantification of nicotine and metabolites, TNE, and nicotine metabolite ratio (NMR) between direct and indirect LCMS methods. DNA from a subset (n=97 smokers) was genotyped for UGT2B10*2 and UGT2B17*2 and the known impact of these variants was evaluated using urinary ratios determined by the direct versus indirect method.
Results
The direct method showed high accuracy (0-9% bias) and precision (3-14% coefficient of variation) with similar distribution of nicotine metabolites to literary estimates and good agreement between the direct and indirect methods for nicotine, cotinine, and 3-hydroxycotinine (ratios 0.99-1.07), but less agreement for their respective glucuronides (ratios 1.16-4.17). The direct method identified urinary 3HC+3HC-GLUC/COT as having the highest concordance with plasma NMR and provided substantially better estimations of the established genetic impact of glucuronidation variants compared to the indirect method.
Conclusions
Direct quantification of nicotine and metabolites is less time-consuming and less costly, and provides accurate estimates of nicotine intake, metabolism rate and the impact of genetic variation in smokers.
Impact
Lower cost and maintenance combined with high accuracy and reproducibility make the direct method ideal for smoking biomarker, NMR and pharmacogenomics studies.
Keywords: Nicotine, Liquid Chromatography, Tandem Mass Spectrometry, Biomarkers, Total Nicotine Equivalents
INTRODUCTION
The risk of tobacco-related diseases increases with smoking dose (1). The best biomarker of dose is determined by the sum of urinary concentrations of nicotine (NIC) and metabolites known as total nicotine equivalents (TNE) (2). TNE, when measured as the molar sum of NIC and eight major metabolites referred to as TNE 9, accounts for approximately 85-90% of the NIC dose (3) and is not significantly affected by individual differences in metabolism, and resulting metabolite levels (4-6), making it the gold standard in estimating tobacco exposure (2, 7). NIC metabolism occurs primarily by CYP2A6-catalyzed C-oxidation into COT. Non C-oxidation pathways of NIC metabolism include glucuronide conjugation, primarily by UGT2B10, into nicotine glucuronide (NIC-GLUC) and N-oxidation, primarily by FMO3, into nicotine N-oxide (NNO). A small amount of nornicotine (NNIC) is also formed as a product of oxidative N-demethylation. COT is further metabolized into 3-hydroxycotinine (3HC) exclusively by CYP2A6 and to cotinine glucuronide (COT-GLUC) primarily by UGT2B10. A small amount of cotinine N-oxide (CNO) and norcotinine (NCOT) are also formed. 3HC is the main metabolite detected in the urine of smokers and is further metabolized into 3HC glucuronide (3HC-GLUC) primarily by UGT2B17. Accuracy in urinary TNE measurement is important in refining risk prediction models for smoking-related diseases that include tobacco exposure, for example, lung cancer (8).
Characterization of the rate of NIC clearance in smokers, which influences dependence, smoking behavior, and response to smoking cessation treatments (9-13), has important epidemiologic and pharmacologic applications. The CYP2A6 enzyme is responsible for ~90% of the metabolism of NIC to COT and 100% of the metabolism of COT to 3HC (14, 15). The plasma-derived ratio of these two nicotine metabolites (3HC/COT), known as the nicotine metabolite ratio (NMR), can be employed in smokers as a phenotypic biomarker of CYP2A6 enzymatic activity and the rate of nicotine metabolism (16). Due to the large contribution of CYP2A6 to NIC clearance, plasma NMR is also a good proxy for total NIC clearance with clinical applications in smoking cessation outcomes (12, 13, 16-18). Other measures of CYP2A6 activity such as urinary measures of NMR (e.g. 3HC+3HC-GLUC/COT, 3HC/COT, and 3HC+3HC-GLUC/COT+COT-GLUC) have also been used to examine smoking phenotypes and tobacco-related disease risk (19, 20).
The ratio of the glucuronide metabolite to the parent compound is used to phenotype glucuronidation enzymes such as UGT2B10 and UGT2B17. A missense mutation in UGT2B10 (UGT2B10*2, rs61750900) reduces NIC and COT glucuronidation as indicated by lower NIC-GLUC/NIC and COT-GLUC/COT ratios (21). Similarly, a loss-of-function variant in UGT2B17 (UGT2B17*2) reduces 3HC glucuronidation resulting in lower 3HC-GLUC/3HC ratio (22). Accurate estimation of these phenotypic ratios is important in assessing the impact of novel genetic variation in UGT2B10 and UGT2B17 on enzymatic activity which in turn alters tobacco constituent metabolism, including tobacco-specific nitrosamines (23, 24). Differential inactivation of tobacco-specific nitrosamines due to different patterns of UGT2B10 and UGT2B17 variation between ethnic groups likely contributes to the disparity in tobacco-related lung cancer risk observed between these populations. Furthermore, although UGT2B10-mediated nicotine glucuronidation generally represents a minor pathway for nicotine metabolism with nicotine glucuronide constituting 3-5% of total nicotine urinary recovery (25, 26), it can increase substantially up to 40% in Asian and African American populations due to higher frequency of reduced and/or null-activity CYP2A6 variants (27). Several LCMS methods are available for determination of nicotine metabolites. Most methods involve enzymatic or chemical de-glucuronidation for quantification of the glucuronide analytes (NIC-GLUC, COT-GLUC, and 3HC-GLUC) where free compounds are measured before (free) and after de-conjugation (total), and the glucuronide metabolites are estimated as the difference between total and free (5, 25, 28-34). Analytical methods allowing for direct quantification of the glucuronides are also available, albeit not widely used (21, 35). Indirect and direct methodologies use essentially identical LCMS conditions; the primary difference between the two methods is that the direct method does not require the de-conjugation step, instead it uses matching deuterated internal standards for each of NIC-GLUC, COT-GLUC and 3HC-GLUC to quantify these metabolites, while the indirect method analyzes free compounds before and after de-conjugation.
In this study, we describe a direct analytical approach to measuring TNE, eliminating the need for the de-conjugation step to estimate NIC-GLUC, COT-GLUC, and 3HC-GLUC concentrations. We then use a series of urine samples from smokers to explore the reproducibility of individual components of the TNE by the direct method. Next, we examine the relationship between plasma and urinary versions of NMR measured by the direct method to identify which urinary NMR best correlates to plasma NMR. Finally, we use the established impact of two commonly genotyped, reduced- (UGT2B10*2, rs61750900) or loss-of function (UGT2B17*2) genetic variants in UGT2B10 and UGT2B17 on glucuronidation ratios to further investigate the utility of the direct method.
MATERIALS AND METHODS
Reference standards, chemicals and reagents
(−)-Nicotine (NIC) hydrogen tartrate salt, (-)-cotinine (COT) and β-glucuronidase type H1 from Helix pomatia, ammonium acetate, ammonium formate, concentrated hydrochloric acid, acetic acid, formic acid, isopropanol and dichloromathane were purchased from Sigma (St. Louis, MO). (±)-Nicotine-d4 (NIC-d4), (±)-cotinine-d3 (COT-d3), trans-3′-hydroxycotinine (3HC), trans-3′-hydroxycotinine-d3 (3HC-d3), (R,S)-nornicotine (NNIC), (R,S)-norcotinine (NCOT), (R,S)-norcotinine-pyridyl-d4 (NCOT-d4), (1′S, 2′S)-nicotine-1′-oxide (NNO), (±)-trans nicotine-N-oxide-methyl-d3 (NNO-d3), (S)-cotinine N-oxide (CNO), (R,S)-cotinine-d3 N-oxide (CNO-d3), nicotine-N-β-D-glucuronide (NIC-GLUC), nicotine-d3 N-β-D-glucuronide (NIC-GLUC-d3), cotinine N-β-D-glucuronide (COT-GLUC), cotinine-d3 N-β-D-glucuronide (COT-GLUC-d3), trans-3′-hydroxycotinine O-β-D-glucuronide (3HC-GLUC-d3) and trans-3′-hydroxycotinine-d3 O-β-D-glucuronide (3HC-GLUC-d3) were obtained from Toronto Research Chemicals (North York, Canada). (R,S)-Nornicotine-d4 (NNIC-d4) was purchased from CDN Isotopes (Pointe-Claire, Canada). Solid phase extraction cartridges (Oasis® MCX (60 mg, 3 ml)) were purchased from Waters (Milford, MA). Methanol was obtained from Caledon Laboratory Chemicals (Georgetown, Canada). Concentrated ammonium hydroxide was purchased from Fischer Scientific (Pittsburg, PA). All reagents and solvents were HPLC grade. Urinary creatinine concentrations were determined using a colorimetric assay according to protocol provided in Creatinine Assay Kit purchased from Sigma (St. Louis, MO) using a SynergyMX Analyzer from BioTek (Winooski, VT).
Calibrator and quality control solutions
Calibrators and quality control (QC) samples were prepared in 0.01 M HCl and spiked into the urine matrix prior to extraction for final concentrations of 1.0 – 1000 ng/ml. For the direct method, blank urine was diluted 1:9 with 0.01 M HCl and 0.100 ml of the mixture was used as urine matrix. The lower 1:9 dilution was used to enhance measurement of both low and high quantity metabolites in one analytical run. For the indirect method the dilution was 1:19 and 0.100 ml of the mixture was used.
Clinical study samples
The blood and urine samples were collected at baseline, in a subset of participants, as part of the Pharmacogenetics of Nicotine Addiction Therapy (PNAT2) clinical trial for smoking cessation treatment (NCT01314001) and stored at −80°C prior to analyses (13). The clinical study was approved by the Institutional Review Boards at the Center for Addiction and Mental Health (Toronto, Canada) where samples were collected and at the University of Toronto (Toronto, Canada) where samples were analyzed. All participants provided written informed consent. Demographic characteristics for participants who provided blood and urine samples for the current study are presented in supplementary material Table-S1. COT and 3HC were assessed by LCMS/MS in blood prior to the current analysis according to a previously described method (36, 37); limits of quantification were <1 ng/ml. NMR measured in whole blood is highly correlated with plasma NMR with a Spearman correlation coefficient of rho=0.93 (36).
Analytical procedures
Direct Method
Using the direct method, urinary concentrations of NIC, COT, 3HC, NIC-GLUC, COT-GLUC, 3HC-GLUC, and NNIC, NCOT, NNO and CNO were assessed during one analytic procedure. Urine samples were diluted 1:9 with 0.01 M HCl. Each diluted urine sample (0.100 ml) was prepared and extracted using a solid-phase extraction procedure adapted from a previously established method (35) which was modified to include 3HC-GLUC; details are provided in supplementary material Table-S2. The re-constituted samples were separated chromatographically using previously published conditions (31, 32). Specifically, an Agilent 1260 HPLC system (Agilent Technologies, Santa Clara, CA) was equipped with a 4.6 mm 150 mm Synergi Polar-RP column (4 mm) (Phenomenex, Torrance, CA). The mobile phase consisted of a 10 mM ammonium acetate/0.1% acetic acid in water (solvent A) and 10 mM ammonium acetate/0.1% acetic acid in methanol (solvent B). The flow rate was 0.7 ml/min. The initial composition was 80% solvent A, changing to 100% solvent B over 6.5 minutes. From 6.5 to 8 minutes, 100% solvent B was maintained and was then changed to 80% solvent A at 8.1 minutes and maintained at this composition until the end of the run; total run time was 13 minutes. The retention times, monitored mass transitions and specific settings for each analyte are provided in supplementary material Table-S3. The linear range, R2, and the limit of quantification (LOQ) were 1–1000 ng/ml, >0.99, and 1 ng/ml for all standards, respectively.
Indirect Method
For the indirect method, as previously described, free concentrations refer to measurements assessed without enzymatic de-conjugation while total levels were assessed after enzymatic de-conjugation. Concentrations of glucuronide-conjugated NIC, COT and 3HC were calculated as the difference between measured total (post de-conjugation) and free (measured without de-conjugation) concentrations for each compound.
Urine samples were diluted 1:19 with 0.01 M HCl prior to analysis. Each urine sample (0.100 ml) was prepared and extracted following a previously described procedure (31, 32, 38); for details see supplementary material Table-S2. The retention times, monitored mass transitions and specific settings for each analyte are provided in supplementary material Table-S3. The linear range, R2, and the limit of quantification (LOQ) were 1–1000 ng/ml, >0.99, and 1 ng/ml for all standards, respectively.
The indirect method detects free compounds only, thus the samples were enzymatically de-conjugated to measure total (free plus glucuronide-conjugate) concentrations of NIC, COT and 3HC. A 0.100 ml aliquot of 1:19 diluted urine was treated with 4000 units of β-glucuronidase in 0.200 ml 0.5 M ammonium acetate solution (pH 5.1). To the mixture, 0.100 ml internals standard solution consisting of 20 ng/ml of NIC-d4, COT-d3, 3HC-d3 and NNO-d3 in 0.01 M HCl, 0.200 ml 0.5M ammonium acetate (pH 5.1), and 0.400 ml HPLC-grade water was added. The samples were incubated for 18 hours (tested from 0-18 hr) at 37°C with shaking and extracted as described (supplementary material Table-S2). All 1000μl was then applied to the solid phase extraction column.
For either method, any samples with analytes detected below the linear range of the calibration curve were re-analyzed using a smaller dilution factor.
Genotyping
In a subset of participants with Caucasian ethnicity (n=97), DNA was extracted from blood by HealthGene Corporation (Vaughan, ON) and genotyped for polymorphisms in UGT2B10 and UGT2B17 genes; analysis was restricted to Caucasians to reduce the potential impact of population stratification. The UGT2B10*2 (rs61750900) polymorphism was determined by real-time PCR using a custom design TaqMan SNP genotyping assay (w1506724762000; Life Technologies). The UGT2B10 rs11726322 genotypes were ascertained from a genome-wide SNP genotyping performed using the Illumina HumanOmniExpressExome-8 v1.2 array (Illumina, San Diego, CA) and imputed using IMPUTE2 software with a quality score of >0.4. The UGT2B17*2 deletion allele was assessed by real-time PCR using the TaqMan gene expression assay (HS00854486_sH; Life Technologies) with copy number reference assay RNase P for internal control (39, 40).
Statistical analysis
Since indirect method does not detect cotinine N-oxide (CNO), this metabolite was excluded from these analyses. We used two definitions of TNE – TNE 6 (6 compounds), the sum of NIC, COT, 3HC, NIC-GLUC, COT-GLUC and 3HC-GLUC and TNE 9 (9 compounds), the sum of TNE 6 plus NNIC, NCOT and NNO. All measurements of analytes were non-normally distributed and were therefore log-transformed for further analyses. Bland–Altman analysis was used to test the level of agreement between the two methods (41). The Bland-Altman ratios were calculated by first taking the difference between the paired measurements by the direct method and the indirect method for each sample and then calculating the average of these differences for all 139 samples. The Bland-Altman range of agreement was defined as the mean bias ± 2 SD. In addition to Pearson correlation coefficients, we used linear regression to determine the strength of associations between the measurements by the two methods and between plasma and urinary NMR and C-oxidation measurements. Mann Whitney U Tests were used to compare within-method effect of UGTB10 genotypes on NIC and COT glucuronidation ratios and Kruskal Wallis Tests were used to assess the within-method differences in 3HC glucuronidation ratios by UGT2B17 genotypes. Linear regression analyses were carried out with GraphPad Prism (version 6.0) software and the Kruskal Wallis and Mann Whitney Tests were performed in IBM SPSS Statistics 24.
RESULTS
Precision and accuracy of the direct method
The direct method showed high accuracy (0-9% bias) and precision (3-14% coefficient of variation, CV) which was tested in three nicotine-free urine samples spiked with each of ten analytes to three different concentrations (10, 100, and 1000 ng/ml) measured over 4-5 days. The precision of the direct method was also tested separately in fifteen smokers’ urine samples measured over 3-4 days with less than 15% coefficient of variation (8-15% CV) (supplementary material Table-S4 and Table-S5). The metabolic profile of nicotine as determined by the direct method is presented in Figure-1; the proportion of nicotine and all nine metabolites as a fraction of all metabolites excreted in urine (i.e. TNE) fell within the expected range in Caucasians (3, 25, 26, 42-44).
Figure-1.

Metabolic profile of nicotine as measured by the direct method. The metabolic profile of nicotine is shown as a pie chart representing the molar amount of each metabolite found in urine when measured by the direct method, expressed as a percentage of the total nicotine equivalents. Also shown are the molar sum of all nicotine metabolites formed by C-oxidation (COT + 3HC + their respective glucuronides, NNIC and NCOT), and nicotine metabolites generated via pathways other than by C-oxidation (NIC GLUC and NNO), and free NIC as a fraction of the total of all nicotine and metabolites (TNE 9) excreted in urine.
Comparison of individual metabolite concentrations, TNE and NMR by the direct and indirect methods
The absolute urinary measurement of each analyte by the direct method is presented in supplementary material Table-S6. Based on Bland-Altman analysis, where a ratio of 1 indicates the least difference between measures, the direct method provided similar measures of NIC, COT, 3HC, and NNO (ratios of 0.99-1.08) (Table-1) compared to when analytes were measured by the indirect method. Correlations between measures of NIC, COT, 3HC, and NNO were high across the two methods (Pearson r=0.88-0.94) (Table-1) as expected, given the similarities of the direct and indirect methods in measuring these free compounds (Table-1).
Table 1.
Bland-Altman analysis of agreement and Pearson correlations of nicotine and metabolites, TNE, and NMR measured in urine according to the direct method and the indirect method
| Ratioa | 95 % CIb | Range of agreementc | Correlation | |
|---|---|---|---|---|
| 1. Nicotine and metabolites (nmol/mg creatinine) | ||||
| NIC | 0.99 | 0.91-1.07 | 0.38 – 2.55 | r=0.92, P<0.0001 |
| COT | 1.07 | 1.01-1.12 | 0.56 – 2.03 | r=0.89, P<0.0001 |
| 3HC | 1.06 | 1.00-1.13 | 0.50 – 2.28 | r=0.94, P<0.0001 |
| NNIC | 1.81 | 1.51-2.18 | 0.20 – 16.7 | r=0.63, P<0.0001 |
| NCOT | 1.37 | 1.23-1.52 | 0.37 – 5.02 | r=0.77, P<0.0001 |
| NNO | 1.08 | 1.00-1.17 | 0.42 – 2.77 | r=0.88, P<0.0001 |
| NIC GLUC | 4.17 | 3.46-5.03 | 0.44 – 39.3 | r=0.57, P<0.0001 |
| COT GLUC | 2.12 | 1.89-2.39 | 0.52 – 8.69 | r=0.85, P<0.0001 |
| 3HC GLUC | 1.16 | 0.97-1.40 | 0.13 – 10.4 | r=0.75, P<0.0001 |
|
| ||||
| 2. Total nicotine equivalents (nmol/mg creatinine) | ||||
| TNE 6 | 1.23 | 1.16-1.31 | 0.59 – 2.58 | r=0.90, P<0.0001 |
| TNE 9 | 1.22 | 1.16-1.29 | 0.62 – 2.43 | r=0.91, P<0.0001 |
|
| ||||
| 3. Nicotine metabolite ratios | ||||
| 3HC+3HC GLUC/COT | 1.01 | 0.96-1.05 | 0.59 – 1.71 | r=0.94, P<0.0001 |
| 3HC/COT | 0.98 | 0.93-1.03 | 0.56 – 1.72 | r=0.93, P<0.0001 |
| 3HC+3HC GLUC/COT+COT GLUC | 0.67 | 0.63-0.71 | 0.32 – 1.42 | r=0.83, P<0.0001 |
The mean difference between log-transformed measures is back-transformed and shown here.
The 95% confidence interval of the mean difference between log-transformed measures is back-transformed and shown here.
The mean difference between log transformed measures ± 2 std. devs is shown here.
We observed moderately lower agreement in 3HC-GLUC measurement between the direct method and the version of the indirect assessment described here. Although Bland-Altman ratio for 3HC-GLUC was close to 1 (ratio of 1.16), the between-method correlation was weaker (Pearson r=0.75) (Table-1). Similarly, we observed lower agreement in NIC-GLUC and COT-GLUC measurements according to Bland-Altman ratios of 4.17 and 2.12 with between method correlations of Pearson r=0.57 and 0.85, respectively. The agreement was lower for NNIC and NCOT as well (ratios of 1.81 and 1.37, respectively) likely due to the difference in use of internal deuterated standards between the two assays and the relatively low concentration of these compounds in smokers’ urine close to the limit of detection for both methods. Figure-2 (A-I) summarizes linear regression analysis for nicotine and metabolites. NIC, COT, and 3HC had relatively high regression coefficients ranging from 0.79 to 0.88. COT-GLUC and 3HC-GLUC exhibited moderate regression coefficient values ranging from 0.57 to 0.71, while NIC-GLUC had low regression coefficient value of 0.33. Together, this suggests good concordance for NIC, COT, and 3HC, but less concordance for NIC-GLUC, COT-GLUC, and 3HC-GLUC between the direct and the version of the indirect method described here.
Figure-2.

Regression analysis of nicotine and metabolites, TNE, and NMR measured by the direct method and the indirect method. Each point on the plots represents an individual measurement of a sample by the direct compared to the indirect method. R2 and P-value shown are from linear regression analysis. All data were log-transformed. Nicotine and metabolites were expressed as ng/ml and TNE was expressed as nmol/mg creatinine.
The direct method provided slightly higher estimations for TNE 6 and TNE 9 compared to the indirect method with Bland-Altman ratios of 1.23 and 1.22, respectively (Table-1). However, TNE 6 and TNE 9 were highly correlated (Pearson r=0.90 and r=0.91, respectively), with high regression coefficients (R2=0.85 and 0.83, respectively) (Figure-2) when measured by the direct compared to the indirect method, suggesting good concordance across methods in providing estimates for urinary total nicotine equivalents.
Evaluation of interference with metabolite detection by β-glucuronidase in the indirect method
To further investigate the apparent loss of NIC-GLUC, COT-GLUC, and 3HC-GLUC measurements using the indirect method, we tested the efficiency of enzymatic de-glucuronidation by incubating blank urine samples spiked with glucuronide standards with β-glucuronidase, and analyzing the samples using the extraction and MS/MS conditions from the direct method to detect the glucuronide concentrations; negligible concentrations of glucuronide compounds remained after 4 hours which did not decrease upon further incubation up to 18 hours (supplementary material Figure-S1).
In order to investigate whether pre-treatment of the samples with β-glucuronidase interferes with accurate quantification of total NIC, total COT, and total 3HC in our version of the indirect method, two additional experiments were conducted in smokers’ urine samples. In the first experiment, the concentrations of glucuronide compounds were tested (using the extraction and MS/MS conditions from the direct method) before and after de-conjugation with β-glucuronidase using matching deuterated internal standards; less than 1% of each of the glucuronide compounds remained following de-conjugation (supplementary material Table-S7).
In the second experiment, the concentrations of total NIC, total COT, and total 3HC were tested (using the extraction and MS/MS conditions from the indirect method) with matching isotopically labeled glucuronides as internal standards that were added to the incubation mixture prior to de-glucuronidation (no deuterated NIC, COT and 3HC was added). Concentrations of total NIC, COT and 3HC were quantified using deuterium-labelled aglycons produced during de-conjugation of the added internal standards. The concentrations measured here were compared to concentrations determined when deuterated aglycons were used as internal standards; the two concentrations agreed suggesting a similar proportion of glucuronide compounds in urine and the labeled glucuronide internal standards are de-conjugated.
Bland-Altman ratios for between-methods comparisons were 1.01 and 0.98 when comparing 3HC+3HC-GLUC/COT (total/free) and 3HC/COT (free/free) ratios, respectively, which is indicative of strong agreement across the two methods. Similarly, both ratios were highly correlated with relatively high regression coefficients (Table-1, Figure-2).
We observed moderately lower agreement in the comparison of urinary 3HC+3HC-GLUC/COT+COT-GLUC (total/total) ratio between the two methods (ratio of 0.67), which still exhibited relatively high correlation and regression coefficients across the two methods (Pearson r=0.83 and R2=0.70) (Table-1, Figure-2).
Comparison of urinary nicotine metabolite ratios (NMR) to plasma NMR
Numerous urinary NMR ratios have been utilized without a clear indication of whether they correlate to plasma NMR and by extension to nicotine clearance. High correlations and regression coefficients were observed between NMR measured in plasma and two of the standard urinary NMR measures, by both methods, with the strongest correlations between plasma NMR and urinary 3HC+3HC-GLUC/COT (total/free) and 3HC/COT (free/free) measured by the direct method (Pearson r=0.75, P<0.0001; Pearson r=0.75, P<0.0001) (Table-1, Figure-3 A-C). The correlation between plasma NMR and urinary 3HC+3HC-GLUC/COT+COT-GLUC was moderately lower (Pearson r=0.61, P<0.0001), suggesting the inclusion of COT-GLUC in the denominator weakened the relationship. Other less widely-used phenotype biomarkers for nicotine C-oxidation including urinary COT/NIC and COT+COT-GLUC/NIC (total COT/NIC) ratios were not correlated with plasma NMR (Pearson r=−0.09 and 0.03, respectively) (Figure-3 D and E). Urinary COT+3HC+COT-GLUC+3HC-GLUC+NCOT/NIC was significantly, albeit weakly, correlated with plasma NMR (Pearson r=0.21) (Figure-3 F).
Figure-3.
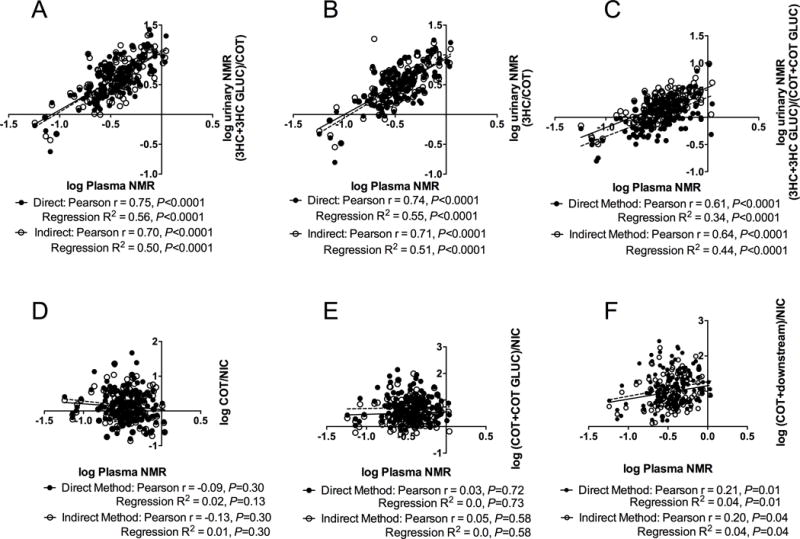
Association of plasma nicotine metabolite ratio with three urinary nicotine metabolite ratios and three urinary C-oxidation ratios. Correlation and linear regression analyses of plasma nicotine metabolite ratio with urinary nicotine metabolite and C-oxidation ratios derived from 2 methodological approaches are shown. Each point on the plot represents an individual measurement of a sample by the direct method (closed circle, solid line) and the indirect method (open circle, dotted line). (F) (COT + downstream)/NIC refers to (COT + 3HC + their respective glucuronide + NCOT)/NIC
Functional impact of UGT2B10 and UGT2B17 polymorphisms on urinary phenotypes
We used ratios of GLUC to parent compound (metabolite ratio) or to TNE (fractional excretion) to examine the impact of established UGT2B10 and UGT2B17 genetic variants known to reduce glucuronidation. Among Caucasians (subset, n=97), UGT2B10*2 (rs61750900) and rs11726322 and UGT2B17*2 allele frequencies were 8%, 12% and 34%, respectively, consistent with previously reported frequencies, and were found to be in Hardy-Weinberg Equilibrium (24). The impact of UGT2B10*2 (rs61750900, reduced-function variant) on NIC and COT glucuronidation was more pronounced with data derived from the direct method than the indirect method (Figure-4 A-D). The same was observed for a second variant in UGT2B10, rs11726322, that is not in linkage disequilibrium with UGT2B10*2 (supplementary material Figure-S2). There was a significant gene-dose decrease in the 3HC glucuronidation ratios with increasing numbers of the UGT2B17*2 deletion alleles using data derived from the direct method, which was not observed with data from the indirect method (Figure-4 E-F).
Figure-4.
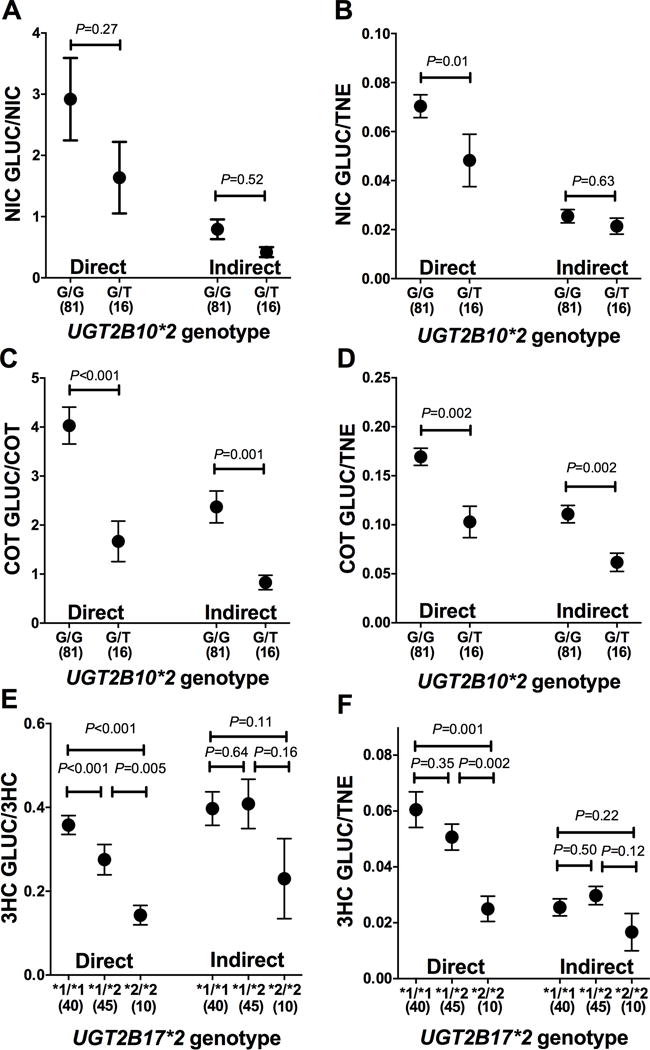
Impact of UGT2B10 and UGT2B17 variation on NIC, COT, and 3HC glucuronidation ratios measured by the direct and indirect methods. UGT2B10*2 genotype and NIC-GLUC/NIC (A), NIC-GLUC/TNE (B), COT-GLUC/COT (C), and COT-GLUC/TNE (D); and UGT2B17*2 genotype and 3HC-GLUC/3HC (E) and 3HC-GLUC/TNE (F) measured by the direct and indirect methods (mean SEM) are shown. These analyses were restricted to the subset of individuals with Caucasian ethnicity (n=97). P-values are for the comparison between genotypes within each method and are generated using Mann Whitney test (A-D) and Kruskal Wallis with Dunn’s Multiple Comparison test (E-F). No individuals with the homozygous variant UGT2B10*2 T/T genotype were identified.
Cost comparisons between direct and indirect analytical methods
The costs differ between the two methods as the direct method requires three additional glucuronide calibration and deuterated standards while the indirect method requires a de-conjugation enzyme, twice the amount of standards and solvents (for the two runs), and twice the usage (i.e. wear and tear) of the columns. The major cost differential, however, lies in the amount of time required to process each sample as the direct method requires only one extraction and LCMS/MS run per sample, while the indirect method requires an additional enzymatic de-conjugation incubation step and as a result over twice the time commitment for sample preparation, extraction and LCMS/MS analysis. Thus, depending on the cost for personnel and equipment maintenance, analysis of one set of samples will cost roughly twice as much with the indirect versus the direct method.
DISCUSSION
Here, we described and validated a direct LCMS approach to quantifying TNE in smokers’ urine. We observed high concordance between plasma NMR and urinary total 3HC/free COT ratio when measured by the direct method. Lastly, we demonstrated the direct method provided substantially better estimations of the established genetic impact of glucuronidation variants (UGT2B10*2, UGT2B17*2) compared to the indirect method.
As many types of indirect and direct methodologies exist across laboratories, the versions of the two methods described here differ from those described previously in their extraction and/or MS/MS conditions. The two methods described here utilized essentially identical LCMS conditions, but differed in the sample preparation process and the internal standards used for each analyte (supplementary material Table-S2). For example, the indirect method used only non-glucuronidated (free) deuterium-labelled internal standards (e.g. NIC-d4) for quantification, while the direct method used a unique deuterium-labelled internal standard for each of the analytes, including the glucuronides (e.g. NIC-GLUC-d4) eliminating the need for a de-conjugation step. Therefore, the sample turn-around time was shortened with the direct method, reducing the risk of the introduction of errors associated with a larger number of steps. Furthermore, considering shorter personnel and equipment time required per sample, the direct method was less expensive compared to the indirect method. The direct method described here is similar to other methods capable of directly detecting and quantifying the glucuronidated nicotine metabolites. It employs a sample extraction procedure using Waters Oasis® MCX SPE cartridges containing a stationary phase shown to bind the three glucuronides of nicotine metabolites, thus allowing for extraction of both free and conjugated metabolites in one step (6, 35, 45, 46). Similar to other direct methods, we acidified urine samples with ammonium formate to pH 2.5 prior to extraction, used methanol with ammonium hydroxide for extraction and employed ESI ionization (6, 35, 46). While a number of different HPLC columns can be used for separation, the metabolites were well resolved on the Phenomenex Synergi Polar RP column. This direct method allows for one-step extraction and quantification of nicotine and its nine metabolites in urine, including 3HC-GLUC that has been missing from previous methods (35) and thus allows for calculation of total nicotine equivalents without the de-glucuronidation step.
The direct method provided slightly higher estimations for TNE 6 and TNE 9 (Bland-Altman ratios of 1.23 and 1.22, respectively), but the correlation and regression coefficients for comparisons between the two methods remained high. TNE is the gold standard for estimating smoking dose and is superior to self-reported cigarettes per day which may be subject to variation in smoking intensity and/or self-report bias (47), and plasma COT which is affected by individual genetic variability in its formation and metabolism (48, 49). Accurate estimates of smoking dose is important for risk prediction models aimed at identifying individuals at higher risk for smoking-related diseases such as lung cancer (8), and for comparisons across ethnicities or sexes, both of which can influence the utility of COT (48). In the case of lung cancer, differences in estimations of the smoking dose in risk prediction algorithms may lead to misidentification of those most likely to develop or die from lung cancer, and therefore, reduce the effectiveness of screening programs.
Plasma NMR was most highly correlated with urinary 3HC+3HC-GLUC/COT (total/free) ratio as measured by the direct method suggesting urinary 3HC+3HC-GLUC/COT could be used to estimate nicotine metabolism rate where plasma is not available. Among urinary measures of NMR, we found good agreement for urinary 3HC+3HC-GLUC/COT (total/free) and 3HC/COT (free/free) between the direct and indirect methods, but lower agreement for 3HC+3HC-GLUC/COT+COT-GLUC (total/total), likely due to the lack of biological plausibility in having a product (versus substrate) in the denominator. Differences in urinary NMR measurements are important to note as the total 3HC/total COT ratio is commonly used in cohort studies as a biomarker for CYP2A6 enzymatic activity and the rate of NIC metabolism (50).
We found the phenotype of both glucuronidation variants in UGT2B10*2 and UGT2B17*2 to be well estimated with the direct method. The indirect method used here may potentially not identify novel variants as impactful, thus, overlooking missing genetic variation and reducing power in genetic association studies.
A strength of this study is that all analyses were run in the same laboratory using the same batches of consumables, calibrators, and internal standards on the same urine samples allowing for a head-to-head comparison of the two analytical methods. A limitation of our analysis is that the comparisons presented apply to the two methods exactly as they are described here and may not be generalizable to other versions of either methods that are run with slightly different sample dilutions, internal standards, sample pre-treatment and extraction, and MS/MS conditions.
In conclusion, the direct method for quantification of nicotine and its metabolites is less time-consuming and costly. It also improves the estimations of intake, metabolism rate, and impact of genetic variation making it a better approach for use in smoking biomarker, NMR and pharmacogenomics studies.
Supplementary Material
Acknowledgments
This work was supported by a Canada Research Chair in Pharmacogenomics (R. Tyndale), National Institutes of Health (NIH) PGRN grant DA020830 (to R. Tyndale and C. Lerman), CIHR (TMH-109787 to R. Tyndale), the Campbell Family Mental Health Research Institute of CAMH, the CAMH Foundation, the Canada Foundation for Innovation (#20289 and #16014 to R. Tyndale and T. George) and the Ontario Ministry of Research and Innovation.
List of Abbreviations
- 3HC-GLUC
3-hydroxycotinine glucuronide
- 3HC
3-hydroxycotinine
- CNO
cotinine N-oxide
- COT-GLUC
cotinine glucuronide
- COT
cotinine
- CV
coefficient of variation
- CYP2A6
cytochrome P450 2A6
- FMO3
Flavin-containing monooxygenase 3
- HPLC
high performance liquid chromatography
- LCMS
liquid chromatography-tandem mass spectrometry
- LOQ
limit of quantification
- NCOT
norcotinine
- NIC-GLUC
nicotine glucuronide
- NIC
nicotine
- NMR
nicotine metabolite ratio
- NNIC
nornicotine
- NNO
nicotine N-oxide
- PCR
polymerase chain reaction
- PNAT2
Pharmacogenetics of Nicotine Addiction Therapy 2
- QC
quality control
- SD
standard deviation
- SNP
single nucleotide polymorphism
- TNE
total nicotine equivalents
- UGT2B10
UDP-glucuronosyltransferase 2 family peptide B10
- UGT2B17
UDP-glucuronosyltransferase 2 family peptide B17
Footnotes
Conflicts of Interest: TPG has consulted for Novartis and Pfizer. RFT has consulted for Apotex and Quinn Emanuel. CL has received study medication and placebo from Pfizer. CL, TPG, and RFT have received unrestricted research funding from Pfizer. The remaining authors declare no conflicts of interest.
References
- 1.Lubin JH, Caporaso N, Wichmann HE, Schaffrath-Rosario A, Alavanja MC. Cigarette smoking and lung cancer: modeling effect modification of total exposure and intensity. Epidemiology. 2007;18(5):639–48. doi: 10.1097/EDE.0b013e31812717fe. [DOI] [PubMed] [Google Scholar]
- 2.Scherer G, Engl J, Urban M, Gilch G, Janket D, Riedel K. Relationship between machine-derived smoke yields and biomarkers in cigarette smokers in Germany. Regulatory Toxicology and Pharmacology. 2007;47(2):171–83. doi: 10.1016/j.yrtph.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Benowitz NL, Jacob P, Fong I, Gupta S. Nicotine metabolic profile in man: comparison of cigarette smoking and transdermal nicotine. Journal of Pharmacology and Experimental Therapeutics. 1994;268(1):296. [PubMed] [Google Scholar]
- 4.Benowitz NL, Dains KM, Dempsey D, Yu L, Jacob P., 3rd Estimation of nicotine dose after low-level exposure using plasma and urine nicotine metabolites. Cancer Epidemiol Biomarkers Prev. 2010;19(5):1160–6. doi: 10.1158/1055-9965.EPI-09-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derby KS, Cuthrell K, Caberto C, Carmella SG, Franke AA, Hecht SS, et al. Nicotine metabolism in three ethnic/racial groups with different risks of lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3526–35. doi: 10.1158/1055-9965.EPI-08-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng S, Kapur S, Sarkar M, Muhammad R, Mendes P, Newland K, et al. Respiratory retention of nicotine and urinary excretion of nicotine and its five major metabolites in adult male smokers. Toxicol Lett. 2007;173(2):101–6. doi: 10.1016/j.toxlet.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Liang Q, Mendes P, Sarkar M. Is 24h nicotine equivalents a surrogate for smoke exposure based on its relationship with other biomarkers of exposure? Biomarkers. 2011;16(2):144–54. doi: 10.3109/1354750X.2010.536257. [DOI] [PubMed] [Google Scholar]
- 8.Ten Haaf K, Jeon J, Tammemagi MC, Han SS, Kong CY, Plevritis SK, et al. Risk prediction models for selection of lung cancer screening candidates: A retrospective validation study. PLoS Med. 2017;14(4) doi: 10.1371/journal.pmed.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ray R, Tyndale RF, Lerman C. Nicotine dependence pharmacogenetics: role of genetic variation in nicotine-metabolizing enzymes. J Neurogenet. 2009;23(3):252–61. doi: 10.1080/01677060802572887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benowitz NL. Clinical pharmacology of nicotine: implications for understanding, preventing, and treating tobacco addiction. Clin Pharmacol Ther. 2008;83(4):531–41. doi: 10.1038/clpt.2008.3. [DOI] [PubMed] [Google Scholar]
- 11.Ho MK, Mwenifumbo JC, Al Koudsi N, Okuyemi KS, Ahluwalia JS, Benowitz NL, et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin Pharmacol Ther. 2009;85(6):635–43. doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerman C, Tyndale R, Patterson F, Wileyto EP, Shields PG, Pinto A, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther. 2006;79(6):600–8. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Lerman C, Schnoll RA, Hawk LW, Jr, Cinciripini P, George TP, Wileyto EP, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Respir Med. 2015;3(2):131–8. doi: 10.1016/S2213-2600(14)70294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messina ES, Tyndale RF, Sellers EM. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 1997;282(3):1608–14. [PubMed] [Google Scholar]
- 15.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996;277(2):1010–5. [PubMed] [Google Scholar]
- 16.Dempsey D, Tutka P, Jacob P, 3rd, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76(1):64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Patterson F, Schnoll RA, Wileyto EP, Pinto A, Epstein LH, Shields PG, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther. 2008;84(3):320–5. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 18.Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacol Biochem Behav. 2009;92(1):6–11. doi: 10.1016/j.pbb.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan JM, Nelson HH, Carmella SG, Wang R, Kuriger-Laber J, Jin A, et al. CYP2A6 genetic polymorphisms and biomarkers of tobacco smoke constituents in relation to risk of lung cancer in the Singapore Chinese Health Study. Carcinogenesis. 2017;38(4):411–8. doi: 10.1093/carcin/bgx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiffman S, Dunbar MS, Benowitz NL. A Comparison of Nicotine Biomarkers and Smoking Patterns in Daily and Non-daily Smokers. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23(7):1264–72. doi: 10.1158/1055-9965.EPI-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Giambrone NE, Jr, Dluzen DF, Muscat JE, Berg A, Gallagher CJ, et al. Glucuronidation genotypes and nicotine metabolic phenotypes: importance of functional UGT2B10 and UGT2B17 polymorphisms. Cancer Res. 2010;70(19):7543–52. doi: 10.1158/0008-5472.CAN-09-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuehl GE, Murphy SE. N-glucuronidation of nicotine and cotinine by human liver microsomes and heterologously expressed UDP-glucuronosyltransferases. Drug Metab Dispos. 2003;31(11):1361–8. doi: 10.1124/dmd.31.11.1361. [DOI] [PubMed] [Google Scholar]
- 23.Chen G, Luo S, Kozlovich S, Lazarus P. Association between Glucuronidation Genotypes and Urinary NNAL Metabolic Phenotypes in Smokers. Cancer Epidemiol Biomarkers Prev. 2016;25(7):1175–84. doi: 10.1158/1055-9965.EPI-15-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wassenaar CA, Conti DV, Das S, Chen P, Cook EH, Ratain MJ, et al. UGT1A and UGT2B genetic variation alters nicotine and nitrosamine glucuronidation in european and african american smokers. Cancer Epidemiol Biomarkers Prev. 2015;24(1):94–104. doi: 10.1158/1055-9965.EPI-14-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benowitz NL, Jacob P., 3rd Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56(5):483–93. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- 26.Byrd GD, Chang KM, Greene JM, deBethizy JD. Evidence for urinary excretion of glucuronide conjugates of nicotine, cotinine, and trans-3′-hydroxycotinine in smokers. Drug Metab Dispos. 1992;20(2):192–7. [PubMed] [Google Scholar]
- 27.Yamanaka H, Nakajima M, Nishimura K, Yoshida R, Fukami T, Katoh M, et al. Metabolic profile of nicotine in subjects whose CYP2A6 gene is deleted. Eur J Pharm Sci. 2004;22(5):419–25. doi: 10.1016/j.ejps.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Murphy SE, Park SS, Thompson EF, Wilkens LR, Patel Y, Stram DO, et al. Nicotine N-glucuronidation relative to N-oxidation and C-oxidation and UGT2B10 genotype in five ethnic/racial groups. Carcinogenesis. 2014;35(11):2526–33. doi: 10.1093/carcin/bgu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecht SS, Carmella SG, Chen M, Koch JFD, Miller AT, Murphy SE, et al. Quantitation of Urinary Metabolites of a Tobacco-specific Lung Carcinogen after Smoking Cessation. Cancer Research. 1999;59(3):590. [PubMed] [Google Scholar]
- 30.Benowitz NL, Jacob P, 3rd, Fong I, Gupta S. Nicotine metabolic profile in man: comparison of cigarette smoking and transdermal nicotine. J Pharmacol Exp Ther. 1994;268(1):296–303. [PubMed] [Google Scholar]
- 31.Jacob P, 3rd, Shulgin AT, Yu L, Benowitz NL. Determination of the nicotine metabolite trans-3′-hydroxycotinine in urine of smokers using gas chromatography with nitrogen-selective detection or selected ion monitoring. J Chromatogr. 1992;583(2):145–54. doi: 10.1016/0378-4347(92)80547-4. [DOI] [PubMed] [Google Scholar]
- 32.Jacob P, 3rd, Yu L, Duan M, Ramos L, Yturralde O, Benowitz NL. Determination of the nicotine metabolites cotinine and trans-3′-hydroxycotinine in biologic fluids of smokers and non-smokers using liquid chromatography-tandem mass spectrometry: biomarkers for tobacco smoke exposure and for phenotyping cytochrome P450 2A6 activity. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879(3–4):267–76. doi: 10.1016/j.jchromb.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu AZ, Zhou Q, Cox LS, Ahluwalia JS, Benowitz NL, Tyndale RF. Variation in trans-3′-hydroxycotinine glucuronidation does not alter the nicotine metabolite ratio or nicotine intake. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benowitz NL, Dains KM, Dempsey D, Wilson M, Jacob P. Racial Differences in the Relationship Between Number of Cigarettes Smoked and Nicotine and Carcinogen Exposure. Nicotine & Tobacco Research. 2011;13(9):772–83. doi: 10.1093/ntr/ntr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller EI, Norris HR, Rollins DE, Tiffany ST, Wilkins DG. A novel validated procedure for the determination of nicotine, eight nicotine metabolites and two minor tobacco alkaloids in human plasma or urine by solid-phase extraction coupled with liquid chromatography-electrospray ionization-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878(9–10):725–37. doi: 10.1016/j.jchromb.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St Helen G, Novalen M, Heitjan DF, Dempsey D, Jacob P, 3rd, Aziziyeh A, et al. Reproducibility of the nicotine metabolite ratio in cigarette smokers. Cancer Epidemiol Biomarkers Prev. 2012;21(7):1105–14. doi: 10.1158/1055-9965.EPI-12-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanner JA, Novalen M, Jatlow P, Huestis MA, Murphy SE, Kaprio J, et al. Nicotine metabolite ratio (3-hydroxycotinine/cotinine) in plasma and urine by different analytical methods and laboratories: implications for clinical implementation. Cancer Epidemiol Biomarkers Prev. 2015;24(8):1239–46. doi: 10.1158/1055-9965.EPI-14-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vieira-Brock PL, Miller EI, Nielsen SM, Fleckenstein AE, Wilkins DG. Simultaneous quantification of nicotine and metabolites in rat brain by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879(30):3465–74. doi: 10.1016/j.jchromb.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38(1):86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 40.Wilson W, 3rd, Pardo-Manuel de Villena F, Lyn-Cook BD, Chatterjee PK, Bell TA, Detwiler DA, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84(4):707–14. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–10. [PubMed] [Google Scholar]
- 42.Neurath GB, Dunger M, Orth D, Pein FG. Trans-3′-hydroxycotinine as a main metabolite in urine of smokers. Int Arch Occup Environ Health. 1987;59(2):199–201. doi: 10.1007/BF00378497. [DOI] [PubMed] [Google Scholar]
- 43.Neurath GB. Aspects of the oxidative metabolism of nicotine. Clin Investig. 1994;72(3):190–5. doi: 10.1007/BF00189309. [DOI] [PubMed] [Google Scholar]
- 44.Gubner NR, Kozar-Konieczna A, Szoltysek-Boldys I, Slodczyk-Mankowska E, Goniewicz J, Sobczak A, et al. Cessation of alcohol consumption decreases rate of nicotine metabolism in male alcohol-dependent smokers. Drug Alcohol Depend. 2016;163:157–64. doi: 10.1016/j.drugalcdep.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rangiah K, Hwang WT, Mesaros C, Vachani A, Blair IA. Nicotine exposure and metabolizer phenotypes from analysis of urinary nicotine and its 15 metabolites by LC-MS. Bioanalysis. 2011;3(7):745–61. doi: 10.4155/bio.11.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piller M, Gilch G, Scherer G, Scherer M. Simple, fast and sensitive LC-MS/MS analysis for the simultaneous quantification of nicotine and 10 of its major metabolites. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;951–952:7–15. doi: 10.1016/j.jchromb.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 47.Klesges RC, Debon M, Ray JW. Are self-reports of smoking rate biased? Evidence from the Second National Health and Nutrition Examination Survey. J Clin Epidemiol. 1995;48(10):1225–33. doi: 10.1016/0895-4356(95)00020-5. [DOI] [PubMed] [Google Scholar]
- 48.Zhu AZ, Renner CC, Hatsukami DK, Swan GE, Lerman C, Benowitz NL, et al. The ability of plasma cotinine to predict nicotine and carcinogen exposure is altered by differences in CYP2A6: the influence of genetics, race, and sex. Cancer Epidemiol Biomarkers Prev. 2013;22(4):708–18. doi: 10.1158/1055-9965.EPI-12-1234-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy SE, Sipe CJ, Choi K, Raddatz LM, Koopmeiners JS, Donny EC, et al. Low cotinine glucuronidation results in higher serum and saliva cotinine in African American compared to White smokers. Cancer Epidemiol Biomarkers Prev. 2017;6(10):1055–9965. doi: 10.1158/1055-9965.EPI-16-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan JM, Nelson HH, Butler LM, Carmella SG, Wang R, Kuriger-Laber JK, et al. Genetic determinants of cytochrome P450 2A6 activity and biomarkers of tobacco smoke exposure in relation to risk of lung cancer development in the Shanghai cohort study. Int J Cancer. 2016;138(9):2161–71. doi: 10.1002/ijc.29963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


