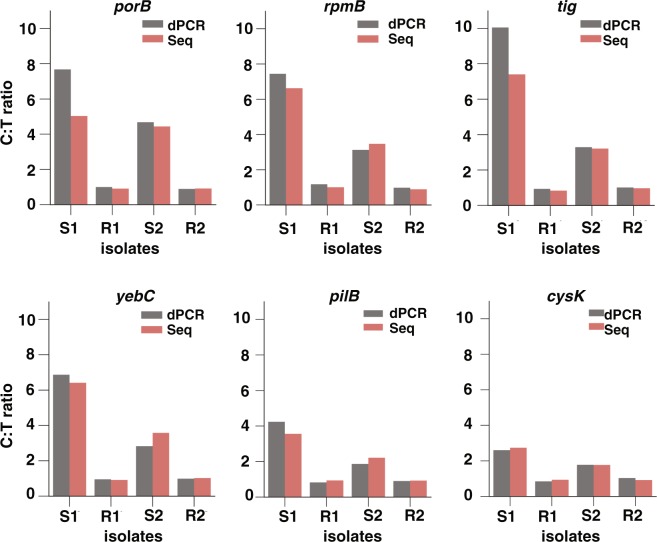
Figure 4.
Validation of the RNA sequencing approach using digital PCR (dPCR) with six candidate markers. Control:treated ratios (C:T ratios) determined by RNA sequencing (red) were validated against C:T ratios measured by dPCR (gray). The dPCR C:T ratios were normalized using ribosomal RNA (rRNA) by dividing the C:T ratio of the candidate marker by the C:T ratio of 16S rRNA. This normalization step is not required for sequencing data because the values are normalized by sequencing depth (see Methods). Markers were validated using two susceptible (S1 and S2) and two resistant (R1 and R2) isolates at 15 min of ciprofloxacin exposure.