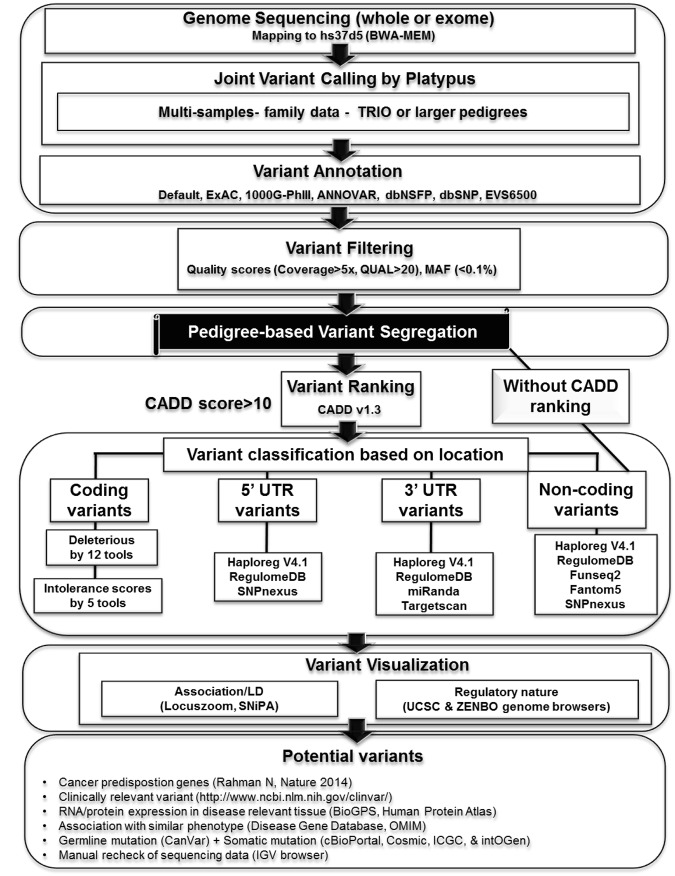
Figure 1.
Summary of familial cancer variant prioritization pipeline version 2 (FCVPPv2). This pipeline uses platypus tool13 for joint variant calling after mapping of the sequencing reads from cases and controls. FCVPPv2 uses several external tools for variant annotation namely ExAC, 1000 Genomes phase III data, ANNOVAR and dbNSFPv3, dbSNP and EVS6500. For candidate variants the variants are filtered using read quality parameters like coverage and quality scores (QUAL) must be >5 and >20, respectively. Minor allele frequency (MAF) must be below 0.1% in the European populations in all used databases. Furthermore, these variants are screened with respective to family-pedigree and this is the most critical step in the germline genomics (shown in black shade). After this step, variants are ranked with the help of CADD v1.320 and any variants with CADD PHRED score of >10 belongs to top 10% for probable functional and deleterious variants in the human genome. These deleterious variants are subsequently divided into 4 different categories based on their locations. The coding variants are considered deleterious based on the consensus from 12 deleteriousness prediction tools and 5 intolerance scores. Variants in the 5′ UTR are considered regulatory based on the Haploreg V4.121, RegulomeDB22 and SNPnexus9 while variants in the 3′ UTR are regulatory if supported by the presence of miRNA binding site using Miranda7 and Targetscan 7.08 tools and additional hints are received from Haploreg V4.121 and RegulomeDB22. For variants in the non-coding segments we combined several state-of- the-art tools such as chromHMM, Segway, FunSeq2 and FANTOM5 data. Non-coding (intergenic and intronic) variants may not always have CADD > 10 even though they will have regulatory implications, so we analyzed all non-coding variants after pedigree segmentation, either with or without CADD > 10. Putative deleterious or regulatory variants are visualized using Locuszoom, SniPA and UCSC genome browser. Potential variants are also checked with sets of additional features, e.g. list of known CPGs1 and clinically relevant variants (ClinVar), expression data and somatic mutations. We also checked the sequencing data of all cases and controls in a particular family for correctness using the IGV browser.