Figure 1. Neuronal expression of CMT2A‐mimetic alleles triggers locomotor defects in flies.
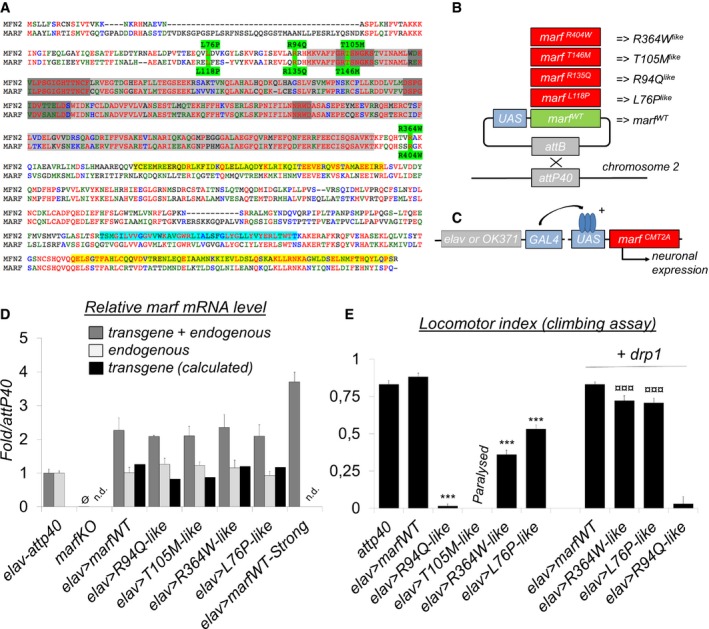
- Sequence alignment of human MFN2 and Drosophila MARF. MFN2 mutations and homologous MARF residues (green), GTP‐binding domain (light gray), GTP‐binding motifs G1 to G4 (dark gray), coiled‐coil/CC domains (yellow), and transmembrane/TM domains (cyan). R94 and T105 are, respectively, located at the edge and within the GTP‐binding domain (position in folded structure see Fig EV1). L76 and R364, that appear distant in the MFN2 linear sequence, are in fact part of the same helix bundle (HB1) as shown in the folded structure (Fig EV1).
- We mutated the amino acids homologous to R94, T105, R364, and L76 in the coding sequence of marf to generate the R404Q, L118P, R135Q, and T146M substitutions (referred here as R364W like, L76P like, R94Q like , and T105M like for simplicity of interpretation), and each marf allele was placed under the transcriptional control of UAS activation sequences. As a control, we similarly generated a wild‐type UAS‐marf construct. The five marf transgenes were inserted at the attP40 insertion site to insure similar genetic background and expression level.
- The expression of the UAS‐marf CMT2A alleles was specifically induced into the neurons of wild‐type flies using the pan‐neuronal driver elav‐GAL4 which targets all central and peripheral neurons or the glutamatergic driver OK371‐GAL4 which facilitates the imaging of motor neurons.
- qRT–PCR performed on larval nervous system extracts showing marf mRNA level relative to control (elav>attp40). “transgene + endogenous”: The amplified sequence is present both in mRNAs encoded by UAS‐marf transgenes and the endogenous marf gene. “endogenous”: amplification of the 3′UTR region that is absent from mRNAs transcribed from UAS‐marf transgenes. The level of “transgene” was calculated (“transgene+endogenous”‐”endogenous”). (mRNA levels ± SD represent the mean value for two independent mRNA extracts each obtained from 12 dissected larval brains). marf KO: knockout of the marf endogenous gene deleting the whole coding sequence and abolishing mRNA expression (Ø : value close to 0) (Appendix Fig S2 for technical details). marf Strong: previously generated UAS‐marf transgene 33 inserted at a different genomic location and leading to higher expression level than the UAS‐marfWT transgene described in this study.
- Result of negative geotaxis test (NGT). Flies were placed into a vertical column and tapped to the bottom to induce escape behavior. After 30 s, the flies reaching the top of the column (N top) and the flies remaining at the bottom (N bot) are counted to calculate a locomotor index (±SEM) defined as: 1/2*(N total + N top − N bot)/N total. Nine races involving three groups of 10 flies for each genotype. Mann–Whitney test results: marfWT versus CMT2A alleles ***P < 0.001. R364W like or L76P like versus same condition + drp1: ¤¤¤P < 0.001. “Paralysis”: NGT not performed on T105M like flies because these animals are completely paralyzed.
