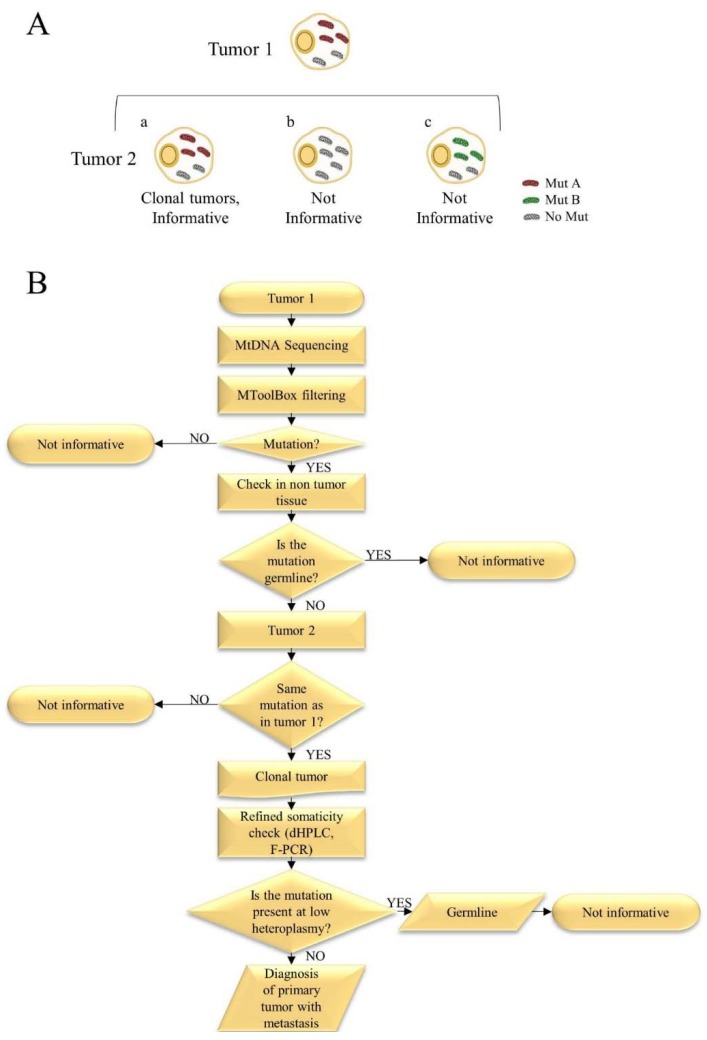
Figure 1.
(A) Schematic representation of mtDNA informativity. Mitochondrial sequencing may be considered as informative only when a tumor-specific mutation is detected in both lesions of the same patient. All the mutations represented in the figure are non-germline mutations. In case (a), a tumor specific mutation occurs in both lesions, indicating clonality. In case (b), Tumor 1 carries a mutation that is not present in Tumor 2 making mtDNA sequencing not informative to infer clonality. In (c), Tumor 1 and 2 carry different mutations and mtDNA sequencing is not informative. In (b and c), it is not possible to rule out a clonal origin of the two masses. (B) The main steps of the mtDNA sequencing workflow: from DNA extraction to clonal diagnosis. mtDNA sequencing is easily and cheaply performed with Sanger method. Analysis of sequences is followed by MToolBox [93] filtering to select non-polymorphic variants. The latter are then checked in the non-tumor tissue or blood to ensure somaticity. If a tumor-specific mutation is detected in Tumor 1, the analysis is shifted to Tumor 2 to identify the same mutation as is Tumor 1. If Tumor 1 and Tumor 2 shared the same mutation a clonal origin may be hypothesized. A refined check of somaticity ought to be performed using more sensitive tools such as dHPLC and F-PCR [95] to rule out a germline mutation present at low heteroplasmic level in non-tumor tissue.