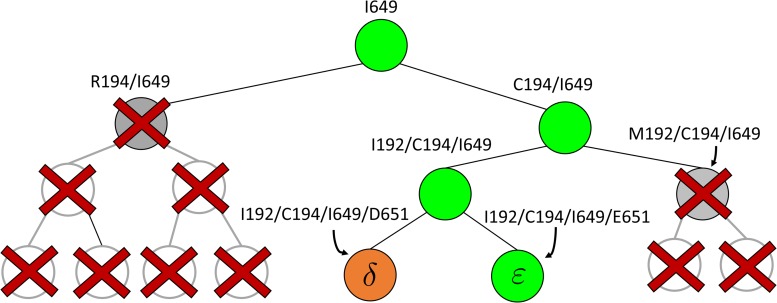
FIG. 2.
BBK* pruning efficiently explores the sequence space. An example design of residues 192 and 194 of the fourth fibronectin F1 module and residues 649 and 651 of a fragment Staphylococcus aureus fibronectin binding protein a-5 is shown (Fig. 1, PDB id: 2RL0). As BBK* searches the sequence space (tree above) its MS bounds provably prunes subtrees from the sequence space. All sequences containing R194/I649 are pruned (red crosses) after computing exactly one MS bound: the bound on the partial sequence R194/I649, which is an upper bound for all sequences containing R194/I649. Sequences containing M192/C194/I649 are pruned (red crosses) after computing only the MS bound for the partial sequence M192/C194/I649. All pruned sequences and partial sequences, shown as empty gray circles, have no additional computation performed. Even though SS bounds are initiated for both I192/C194/I649/E651 and I192/C194/I649/D651, the latter is pruned after computing a mer e 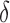 -approximation bound (orange leaf node), which is cheaper, and not as tight as a
-approximation bound (orange leaf node), which is cheaper, and not as tight as a 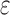 -approximate bound. A provable
-approximate bound. A provable 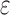 -approximation bound (green leaf node) is computed for only the optimal sequence, I192/C194/I649/E651. In contrast, SS methods compute separate
-approximation bound (green leaf node) is computed for only the optimal sequence, I192/C194/I649/E651. In contrast, SS methods compute separate 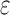 -approximate bounds (which are expensive) for all eight possible sequences, shown as leaf nodes in the tree.
-approximate bounds (which are expensive) for all eight possible sequences, shown as leaf nodes in the tree.