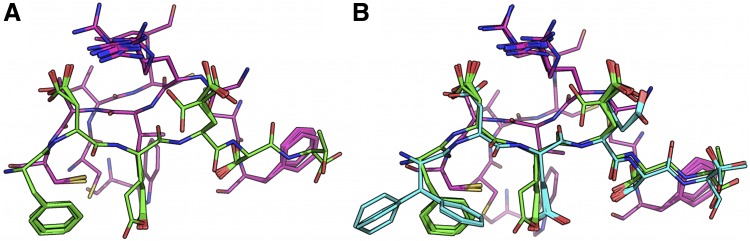
FIG. 5.
BBK* efficiently handles coupled continuous side-chain and local backbone flexibility. Selected residues from ensembles, computed by BBK*, of human fibronectin F1 modules 4–5 (magenta) in complex with a fragment of Staphylococcus aureus fibronectin binding protein A 5 [FNBPA-5, PDB id: 2RL0, (Lower et al., 2011)]. The design space consisted of the wild-type sequence and either 15 or 25 single amino acid mutants. (A) Ensemble of the wild-type sequence based on the original crystal structure. The design used a fixed FNBPA-5 backbone (green) and continuous side-chain flexibility. (B) Ensemble of the wild-type sequence using two backbones: the original FNBPA-5 backbone (green) and a second backbone (PDB id: 2RKY, cyan) with RMSD 1.3 Å from the original [found using the MASTER program (Zhou and Grigoryan, 2015)]. The sequence rankings [by  score, Eq. (2)] from the fixed and flexible backbone models had Spearman correlation coefficients of
score, Eq. (2)] from the fixed and flexible backbone models had Spearman correlation coefficients of  = 0.53 and
= 0.53 and  = 0.82 in the 15 and 25 mutant designs, respectively. This shows that the flexible backbone model favors binding in very different sequences than the fixed backbone model does.
= 0.82 in the 15 and 25 mutant designs, respectively. This shows that the flexible backbone model favors binding in very different sequences than the fixed backbone model does.