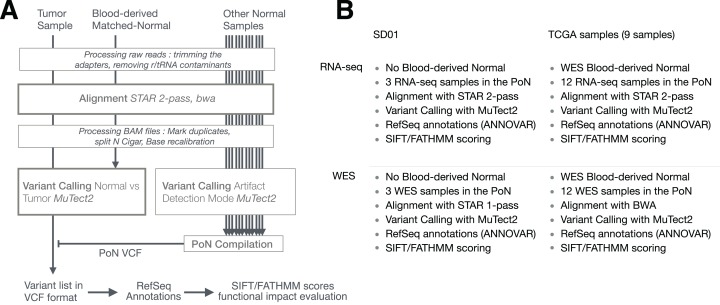
Figure 1. Pipeline used to detect RNA-seq variants.
(A) Principal steps in the pipeline used to identify and annotate somatic mutations. Mutation calling was done for each paired tumor sample/matched-normal. An RNA-seq-specific panel-of-normals (PoN) and a WES-specific PoN were generated. (B) Distinction between pipelines and their associated methodologies for SD01 and TCGA samples, and the difference between RNA-seq and WES pipeline used in this study.