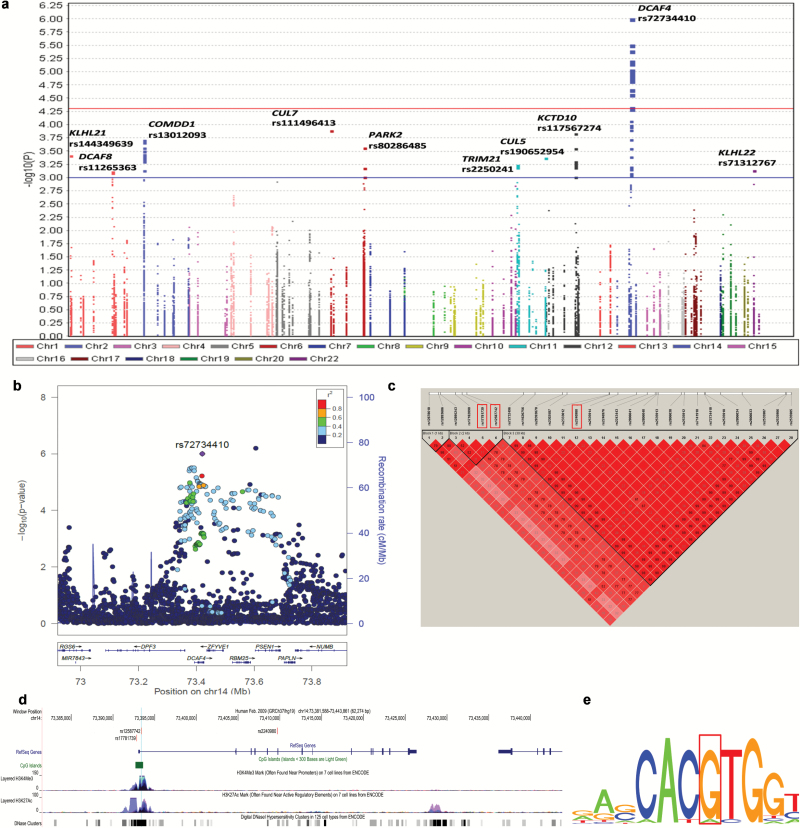
Figure 2.
Association results and functional prediction of SNPs in 115 Cullin-ring ligase encoding genes. (a) Manhattan plot of the overall results. There were 84 SNPs on 10 CRLs genes with nominal P < 0.001 and 28 of them were on the DCAF4 gene with false discovery rate (FDR) < 0.05. The x-axis indicated the chromosome number and the y-axis showed the association P values with lung cancer risk (as –log10 P values). The horizontal blue line represents P values of 0.001 while the red line indicated the FDR threshold 0.05. (b) Regional association plot, which demonstrated that the linkage disequilibrium (LD) between the top SNP rs72734410 on DCAF4 and other SNPs in the region of 500kb up- or downstream of the top SNP. (c) Pair-wise LD plot between the 28 SNPs in DCAF4 with FDR < 0.05. Based on it, two tag SNPs (rs17781739 and rs2240980) together with one functional SNP rs12587742 were chosen for further analysis. (d) Locations and functional prediction of the three selected SNPs. Two SNPs (rs17781739 and rs12587742) are located within one CpG island and presented strong signals of active enhancer and promoter functions (indicated by DNase hypersensitivity, histone modification H3K27 acetylation and H3K4 methylation, respectively). (e) Position weight matrix (PWM) based Sequence Logo, which showed rs12587742 is located on the c-MYC motif.