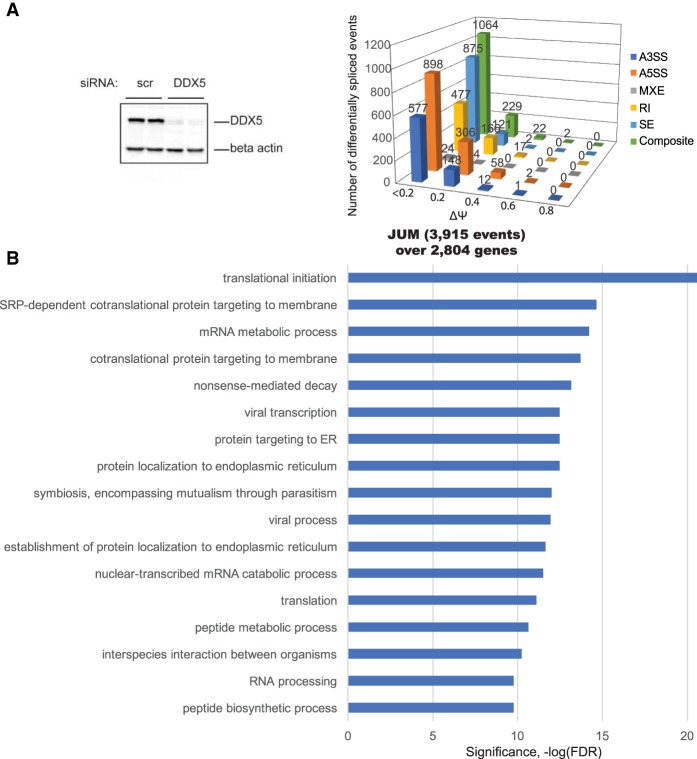
Figure 2.
The RNA helicase DDX controls alternative pre-mRNA splicing of thousands of target RNAs. (A, left panel) K562 cells were transfected with either nonspecific control siRNA oligos (scr si) or DDX5 duplex siRNA oligos. After a second round of siRNA transfection, the cells were harvested for RNA isolation or protein lysates. Protein lysates were immunoblotted with DDX5 antibody to detect the efficiency of siRNA-mediated knockdown at the protein level. (Right panel) Detection of changes in AS events upon DDX5 knockdown in human K562 cells using JUM. JUM analysis revealed 3915 AS events whose splicing patterns were significantly altered in DDX5 RNAi knockdown samples versus the control scrambled siRNA samples, covering 2804 genes (FDR, P < 0.05). (B) GO enrichment analysis of 2804 DDX5 splicing target genes from JUM splicing analysis. Categories of related genes are listed at the left, and the enrichment significance (−log FDR) is indicated along the X-axis.