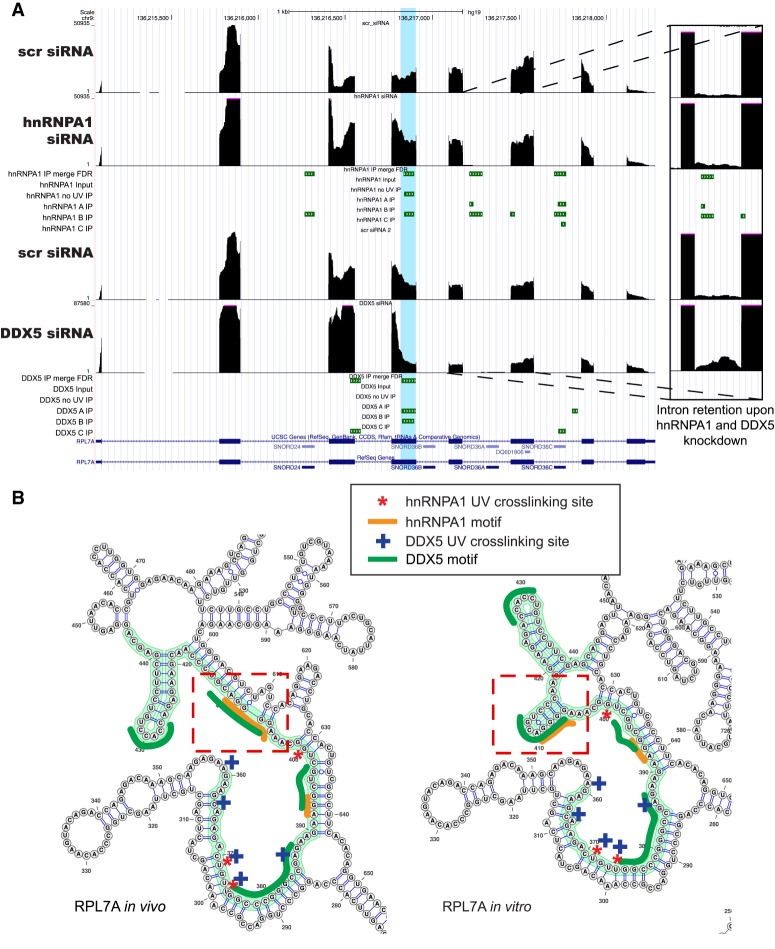
Figure 6.
Alternative pre-mRNA splicing of RPL7A mRNA is regulated by hnRNPA1 and DDX5. (A) Genome browser shot of the RPL7A gene region with RNA-seq data upon hnRNPA1 knockdown (top) and DDX5 knockdown (bottom), with corresponding nuclear CLIP peak cluster regions shown below. (B) icSHAPE-constrained in vivo and in vitro secondary structures for the RPL7A RNA. The nuclear eCLIP cluster/peak region is highlighted in green, the hnRNPA1 cross-link sites are marked with red asterisks, and the DDX5 cross-link sites are marked with blue plus signs. The dotted red rectangles indicate regions of the RNA with secondary structural changes between the in vivo and in vitro icSHAPE constraints. The enriched hnRNPA1 motifs are indicated by orange lines, and the enriched GC-rich motifs for DDX5 is indicated by dark-green lines.