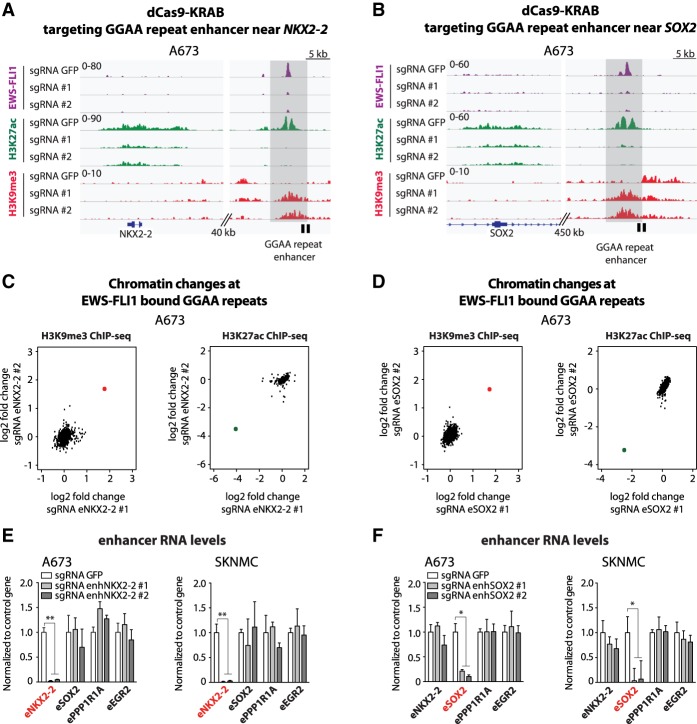
Figure 2.
Site-specific GGAA repeat enhancer silencing using dCas9-KRAB. (A,B) ChIP-seq tracks of FLI1 (EWS-FLI1), H3K27ac, and H3K9me3 signals over the NKX2-2 (A) and SOX2 (B) loci in A673 cells expressing dCas9-KRAB and a sgRNA targeting either the GFP control sequence or GGAA repeat enhancers activated by EWS-FLI1 in Ewing sarcoma. Target regions are shown in light gray, and locations of sgRNA-binding sites are illustrated by a black box. (C,D) Scatter plots show the H3K9me3 and H3K27ac changes observed by ChIP-seq at EWS-FLI1-bound GGAA repeat enhancers (n = 812) in A673 cells lentivirally induced with dCas9-KRAB and sgRNAs targeting enhancers near NKX2-2 (C) or SOX2 (D). A red dot for H3K9me3 and a green dot for H3K27ac show the positions of the targeted enhancers in each experiment. Signals were calculated over a 5-kb window centered on EWS-FLI1-bound GGAA repeats. (E,F) RT-qPCR shows the specific decreases of eRNA levels at targeted EWS-FLI1-bound GGAA repeat enhancers near NKX2-2 (E) or SOX2 (F) in Ewing sarcoma cell lines (A673 and SKNMC). Cells were collected 7 or 8 d after lentiviral transduction. (*) P-value < 0.05; (**) P-value < 0.01. See also Supplemental Figures S3 and S4A.