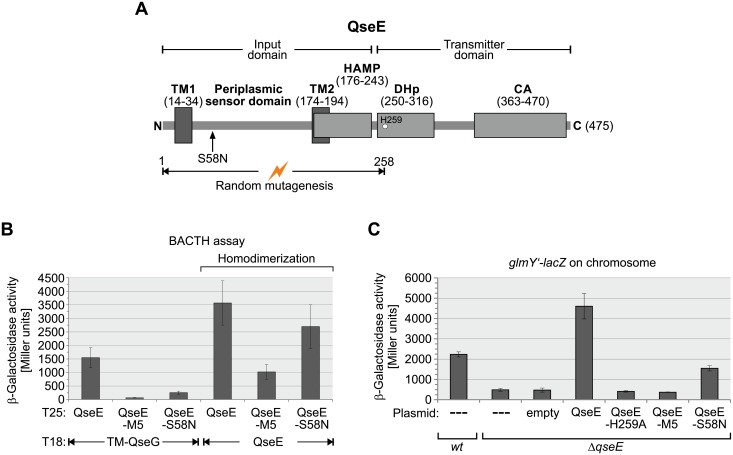
Fig 8. Mutations in the QseE N-terminus impairing interaction with QseG concomitantly decrease QseE activity.
A. Schematic representation of the domain architecture of sensor kinase QseE. Amino acid residues encompassing the respective domains are given in parenthesis and the phosphorylated histidine residue H259 is depicted by a circle. Positions are according to the EcoCyc database [6]. The HAMP domain has been predicted by Pfam [68]. The sequence coding for amino acid residues 1–258 was randomly mutagenized and the resulting QseE mutant library was phenotypically screened for loss of interaction with QseG in the context of BACTH. The position of the thereby identified S58N substitution is indicated with an arrow. B. Quantitative BACTH analysis of the interaction potential of T25-QseE variants identified in the screen for loss of interaction with T18-TM-QseG. The following plasmid combinations were tested in reporter strain BTH101 (left to right): pYG242/pYG199; pYG242/pYG199_TM1; pYG242/pYG199_1.6; pYG246/pYG199; pYG246/pYG199_TM1; pYG246/pYG199_1.6. C. Complementation analysis assessing the ability of QseE variants to activate transcription from promoter PglmY. The following plasmids encoding the proteins under Ptac control were introduced into the ΔqseE mutant Z970 carrying a glmY’-lacZ reporter fusion and the β-galactosidase activities were determined: pKESK23 (empty vector, column 3), pYG221 (wt-QseE, column 4), pYG221-H259A (QseE-H259A, column 5), pYG221-TM1 (QseE-M5, column 6), pYG221-S58N (QseE-S58N, column 7). As controls, un-transformed wild-type (Z197) and ΔqseE (Z970) strains were used (first two columns).