Fig. 2.
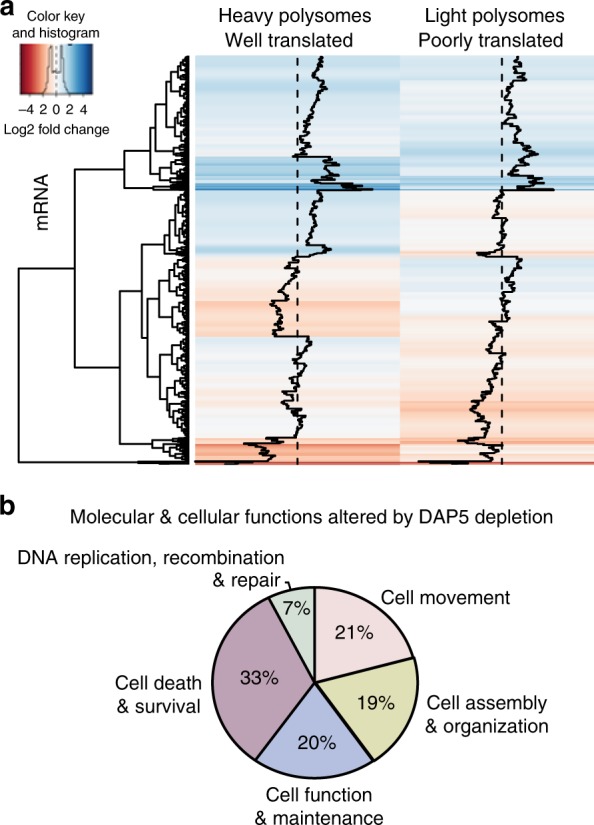
Transcriptomic and translatomic analysis of non-silenced and DAP5-silenced MDA-MB-231 cells. Total mRNA; light polysome, poorly translated mRNA (2–3 ribosome fraction); and heavy polysome, well-translated mRNA (≥4 ribosome fraction) were subjected to RNAseq. Average of two independent complete studies shown. a Heat map of RNAseq-mRNAs dependent on DAP5 for well-translated fractions. The black lines indicate the average fold change per gene across the dataset. Polysome mRNAs most strongly reduced with DAP5 silencing can be found in Supplementary Data 1 for source data. b Top predicted cellular functions affected with DAP5 silencing determined by Ingenuity Pathway Analysis (IPA)
