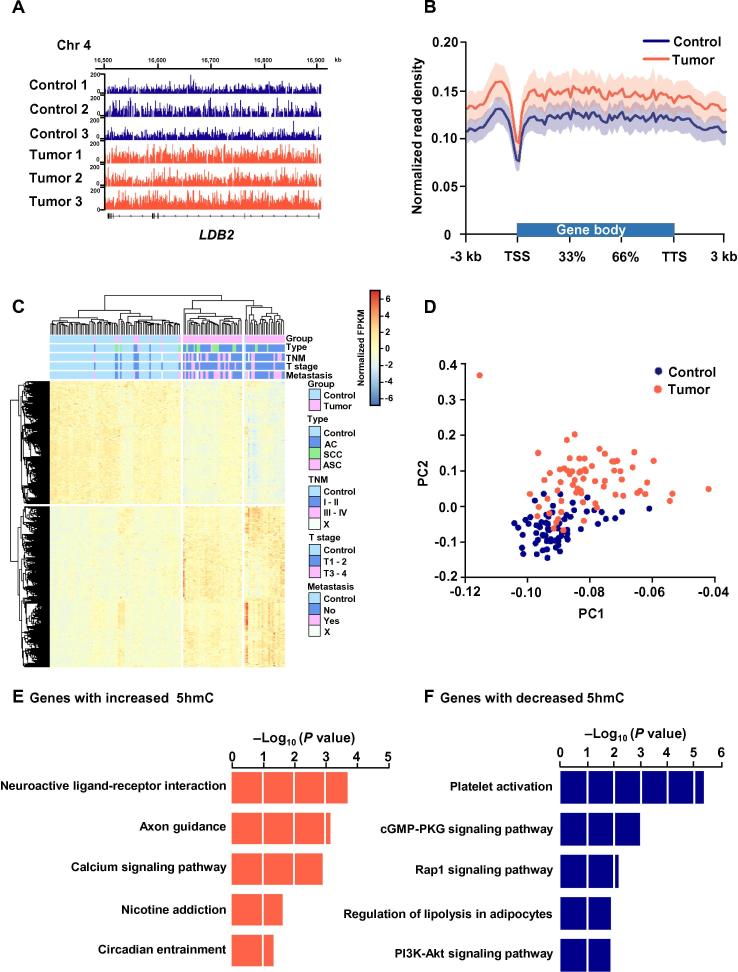
Figure 4.
Identified DhMGs as candidate biomarkers to distinguish blood samples from healthy controls and NSCLC patients
A. Genome browser view of the cell-free 5hmC distribution in LDB2 (one example of differentially-methylated genes) loci in control and tumor samples. The scale represents the rage of normalized read count. B. Differentially methylated metagene profiles of cell-free 5hmC in control and tumor samples. Shaded area indicates the upper and lower quartiles. C. Heatmap of 2459 DhMGs in control and tumor samples. Hierarchical clustering was performed across genes and samples. D. PCA plot of DhMGs FPKM from 67 control and 66 tumor samples. E. KEGG enrichment analysis of genes with significant 5hmC increase in tumor samples. F. KEGG enrichment analysis of genes with significant 5hmC decrease in tumor samples. LDB2, LIM domain binding 2; AC, adenocarcinoma; SCC, squamous cell carcinoma; ASC, adenosquamous carcinoma; DhMG, hyper-hydroxymethylation among the differential gene. X means data not available for classification of TNM stages or metastasis status.