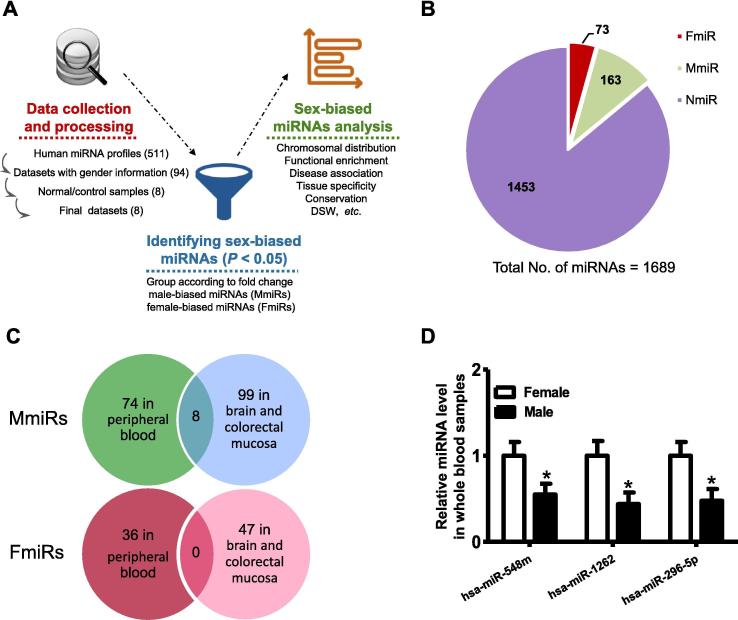
Figure 1.
The overview of the computational framework and the identified the sex-biased miRNAs
A. The overall computational framework of this work. First, human miRNA expression datasets with gender-labeled normal samples were curated from the GEO database. Second, the sex-biased miRNAs across different tissues were screened using Wilcoxon’s test comparing the expression levels between male and female samples (P < 0.05). Finally, various characteristics of these miRNAs were analyzed subsequently using different computational pipelines. B. Fraction of the sex-biased miRNAs in the dataset curated in the current study. C. The overlap of FmiRs and MmiRs between peripheral blood and the other two non-blood tissues, i.e., brain and colorectal mucosa in this study. D. RT-PCR validation of sex-biased miRNA expression in whole blood samples. The expression levels of selected miRNAs were analyzed using t-test in independently collected whole blood samples of males and females as described in the experimental procedures. The RT-PCR assay was performed on the same batch of blood sample. *indicates significant difference in miRNA expression between male and female samples (P < 0.05). N = 8–10. FmiR, female-biased miRNA; MmiR, male-biased miRNA; DSW, disease spectrum width, which is defined as the number of diseases associated with a given miRNA gene divided by the total number of diseases associated with any miRNA genes.